From genotype to phenotype with 1,086 near telomere-to-telomere yeast genomes
Article meta
Article Date: 15 October 2025
Article URL: https://www.nature.com/articles/s41586-025-09637-0
Image: Figure 1
Summary
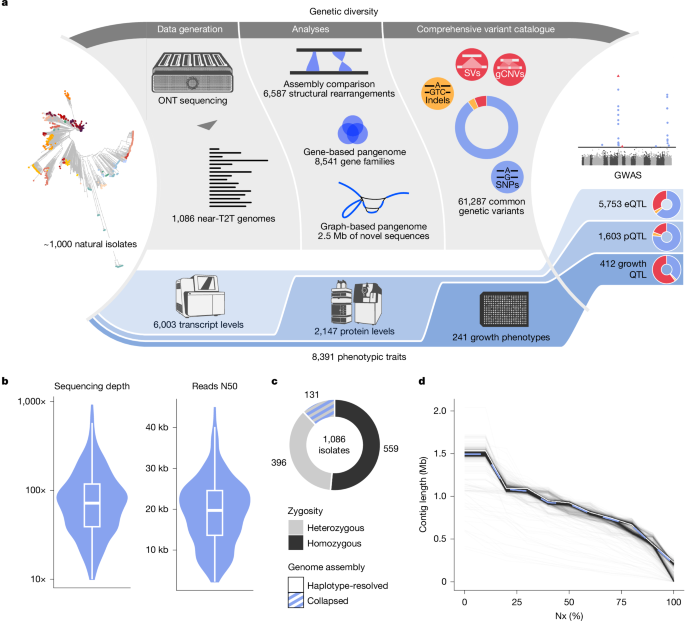
The authors generated near telomere-to-telomere assemblies for 1,086 natural Saccharomyces cerevisiae isolates using long-read sequencing and a hybrid assembly pipeline, producing 1,482 high-quality assemblies. They built both a gene-based pangenome (8,541 gene families, 2,199 non-reference) and a graph pangenome, and produced a comprehensive structural-variant (SV) catalogue (6,587 unique SV events; 262,629 redundant calls across isolates). By integrating these genomic resources with 8,391 molecular and organismal traits they ran GWAS across SNPs, indels and SVs, finding that SVs are disproportionately associated with phenotypic variation and show greater pleiotropy—especially for organismal traits. The graph pangenome added ~2.5 Mb of non-reference sequence and improved genotyping and heritability estimates. Overall, the work demonstrates that an exhaustive, assembly-based pangenome and SV atlas is essential to resolve genotype–phenotype relationships.
Key Points
- Near telomere-to-telomere assemblies were produced for 1,086 S. cerevisiae isolates (1,482 assemblies total), with high contiguity and completeness close to the reference genome.
- A species-wide SV catalogue: 6,587 unique SV events (PAVs, CNVs, inversions, translocations) validated at high rate; SVs span ~27.3 Mb (excluding translocations).
- SVs are enriched in subtelomeric regions and are often linked to Ty transposable elements and repetitive sequences; 46 SV hotspots were identified, some clade-specific and some fragile-region driven.
- Gene-based pangenome: 8,541 gene families (5,047 core; 3,494 accessory), with 2,199 genes absent from the reference—many introgressed or horizontally transferred.
- Including SVs and indels raises trait heritability estimates by ~14.3% on average; a graph pangenome raised heritability by ~10% across >8k traits.
- GWAS found 4,564 QTL (3,471 SNP-QTL, 230 indel-QTL, 863 SV-QTL); SVs are enriched among QTL and show higher pleiotropy (average 2.82 traits per SV-QTL vs 1.45 for SNP-QTL).
- SV types differ in effect: deletions and CNVs show more QTL and larger effects than insertions; indels have the largest average effect size but lower pleiotropy.
- The Minigraph-Cactus graph encodes millions of variant snarls and captures multiallelic and subtelomeric complexity, facilitating genotyping across collections.
Context and relevance
This paper addresses a major gap in population genomics: large, assembly-based SV catalogues across an entire eukaryotic species. While many GWAS focus on SNPs, this study shows that SVs are a major source of phenotypic variation and pleiotropy, especially for organismal (growth) traits. The approach—combining high-coverage long reads, haplotype-resolved assemblies, gene-based pangenomes and graph genotyping—demonstrates a scalable framework for other eukaryotes (including crops and humans) where SVs likely explain part of the missing heritability.
Author style
Punchy: breadth and depth. The paper isn’t just another yeast sequencing project—it’s a species-wide, near-complete map of structural and gene-content variation tied directly to thousands of traits. If you care about how big variants shape phenotypes or about building realistic pangenomes, the analyses and resources here are highly consequential.
Why should I read this
Short and blunt: because these folks actually assembled >1k near-T2T yeast genomes and then used that to show SVs punch above their weight for real traits. If you want evidence that structural variation, gene presence/absence and graph pangenomes matter for trait mapping (and how to do the heavy lifting), this saves you reading a stack of separate papers — they did the grunt work and mapped it back to phenotypes.