Hijacking a bacterial ABC transporter for genetic code expansion
Article Date: 15 October 2025
Article URL: https://www.nature.com/articles/s41586-025-09576-w
Article Image: https://media.springernature.com/lw685/springer-static/image/art%3A10.1038%2Fs41586-025-09576-w/MediaObjects/41586_2025_9576_Fig1_HTML.png
Summary
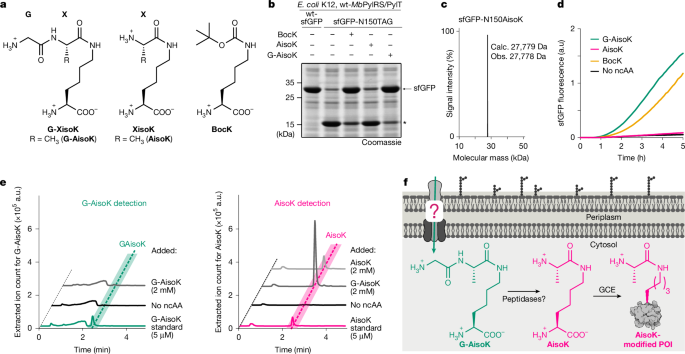
This paper describes a strategy to overcome a major bottleneck in genetic code expansion (GCE): poor intracellular availability of non‑canonical amino acids (ncAAs). The authors design isopeptide-linked tripeptide “propeptides” (Z‑XisoK / G‑XisoK) that are actively imported into Escherichia coli via the oligopeptide permease (Opp) ABC transporter, processed inside the cell by peptidases, and thereby yield high intracellular concentrations of otherwise poorly permeable ncAAs. They solved the OppA–G‑SisoK crystal structure, used FACS‑based directed evolution to reprogramme OppA for selectivity in peptide-rich media, and produced engineered strains (IsoK12, K12‑Z1, K12‑Z2) that enable efficient single and multi‑site incorporation of a broad panel of ncAAs — including previously intractable post‑translational‑modification mimics, bioorthogonal handles and crosslinkers. They also show dual ncAA incorporation from a single tripeptide and demonstrate scalability and improved yields in rich media.
Key Points
- Isopeptide-linked tripeptides (G‑XisoK / Z‑XisoK) act as Trojan horses to deliver ncAAs into E. coli via the Opp ABC transporter.
- Active Opp‑mediated import followed by intracellular peptidase cleavage raises intracellular ncAA concentrations several‑fold compared with direct ncAA supplementation.
- Crystal structure of OppA bound to G‑SisoK reveals that recognition relies on backbone and termini interactions, enabling tolerance for diverse side chains.
- Directed evolution (FACS screen) produced OppA variants (OppA‑iso, OppA‑Z1, OppA‑Z2) and engineered strains (IsoK12, K12‑Z1/Z2) that import tripeptides efficiently in peptide‑rich media, reducing required ncAA concentrations ≈10×.
- The platform enabled efficient incorporation of 11 previously inaccessible XisoK ncAAs, multi‑site suppression (up to triple TAGs), dual ncAA incorporation from a single tripeptide, and production yields comparable with wild‑type protein expression.
- Tripeptides are readily synthesised by solid‑phase peptide synthesis, making the approach scalable and broadly applicable.
Content summary
Genetic code expansion allows site‑specific insertion of ncAAs into proteins, but practical use is limited by low yields and poor uptake of many ncAAs. The authors found that isopeptide‑linked tripeptides (G‑AisoK as an example) are actively transported into E. coli via the Opp oligopeptide ABC transporter and then cleaved by endogenous aminopeptidases (pepN / pepA), releasing the desired ncAA (AisoK) inside the cytosol. This mechanism results in intracellular accumulation of the ncAA at much higher levels than direct supplementation.
They solved the OppA–G‑SisoK X‑ray structure (PDB 9RD1) showing binding driven by backbone and terminal interactions rather than side‑chain specificity, which explains the broad substrate tolerance. Using FACS‑based selection of OppA libraries, the team evolved variants that preferentially bind G‑XisoK tripeptides over competing peptides in nutrient‑rich media. Genomic integration of evolved oppA variants produced strains (IsoK12, K12‑Z1, K12‑Z2) that restore high incorporation efficiencies in 2‑YT and other rich media, enabling higher biomass and scalable production.
Applying the approach, the authors demonstrated efficient encoding of diverse XisoK side chains (bioorthogonal alkynes, diazirines, chloroalanine, PisoK, CisoK and PTM mimics). They validated labelling, photocrosslinking and proximity crosslinking in multiple test proteins and produced milligram quantities of ncAA‑bearing proteins with yields matching wild‑type. They also designed Z‑XisoK tripeptides to deliver otherwise cell‑impermeable Z residues (including bulky or negatively charged ncAAs) and evolved OppA variants to accept these, broadening the chemical space available to GCE. Lastly, they showed dual ncAA incorporation (orthogonal PylRS/tRNA pairs) after uptake and intracellular cleavage of a single tripeptide bearing two different ncAAs.
Context and relevance
Why this matters: many labs use GCE to probe protein function, create conjugates and build modified therapeutics, but uptake and cost of ncAAs limit routine use. This work shifts the bottleneck away from extensive aaRS engineering and expensive high‑concentration supplementation towards engineered import. By turning a native ABC transporter into a programmable import system, the study offers a modular, scalable solution that is immediately relevant to researchers working on protein labelling, PTM studies, protein–protein interactions, and industrial production of modified proteins.
It aligns with broader trends in synthetic biology: decoupling substrate delivery from translation machinery, engineering transporters for non‑natural substrates, and making GCE more practical and economical for larger‑scale applications.
Why should I read this?
If you mess with modified proteins, this is proper useful — the authors cracked a practical way to get stubborn ncAAs into E. coli without buying buckets of exotic amino acids or reinventing aaRSs. It means higher yields, lower reagent cost, and straight‑forward scale‑up. Read it if you want to start making labelled, crosslinked or PTM‑bearing proteins reliably and at scale.
Author style
Punchy: the paper’s practical bent is emphasised throughout — they don’t just demonstrate a clever chemistry trick, they build and evolve strains, solve structure, validate multiple applications and show scalability. For those in protein engineering or biotech, the details (OppA mutations, strain construction, tripeptide designs) are worth diving into.