Mapping Plasmodium transitions and interactions in the Anopheles female
Article Date = 22 October 2025
Article URL = https://www.nature.com/articles/s41586-025-09653-0
Article Title = Mapping Plasmodium transitions and interactions in the Anopheles female
Article Image = https://media.springernature.com/lw685/springer-static/image/art%3A10.1038%2Fs41586-025-09653-0/MediaObjects/41586_2025_9653_Fig1_HTML.png
Summary
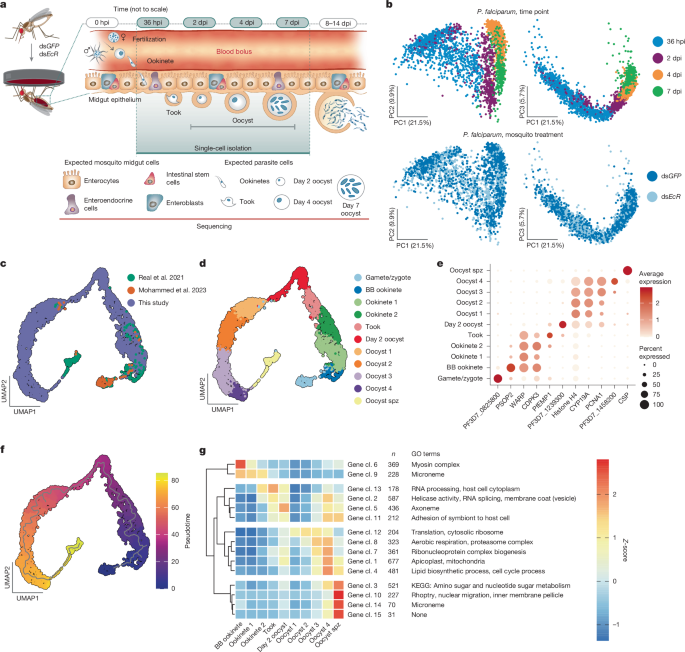
This study uses dual single-cell RNA sequencing of both Plasmodium falciparum parasites and Anopheles midgut cells across key time points (36 hpi, 2 dpi, 4 dpi, 7 dpi) to create a high-resolution atlas of parasite midgut stages. The authors profiled 3,495 parasites, integrated their data with prior datasets to define 11 parasite clusters spanning gametes to segmenting oocysts, and identified gene-expression programmes that drive ookinete invasion, oocyst growth and the onset of sporozoite formation. Functional follow-up in mosquitoes validated several parasite targets (PfATP4, PfLRS, and PfSIP2) as essential for oocyst development or hepatocyte invasion. Dual-read analysis and imaging revealed a conserved preferential interaction between parasites and midgut progenitor cells (ISC/EB) during epithelial traversal, and late oocysts associate closely with visceral muscles.
Key Points
- Dual scRNA-seq of parasites and midgut cells across 36 hpi–7 dpi yielded a complete midgut-stage atlas with 11 parasite clusters and pseudotime trajectory aligning expected transitions.
- Gene-cluster analysis revealed four major expression branches: ookinete maturation/motility, ookinete→oocyst surface remodelling, oocyst growth metabolism/cell cycle, and sporozoite segmentation/invasion machinery.
- Functional validation: inhibiting PfATP4 (cipargamin) or PfLRS (MMV670325) in mosquitoes reduced oocyst growth and largely blocked sporozoite formation; other inhibitors had stage-specific effects.
- Conditional knockdown of PfSIP2 in parasites impairs sporozoite invasion of primary human hepatocytes, linking oocyst-stage transcription factor expression to mammalian infectivity.
- Parasites preferentially interact with intestinal stem cell/enteroblast (ISC/EB) progenitor cells during midgut traversal; late oocysts are physically associated with visceral muscle fibres.
Content summary
The authors enriched parasite cells from infected Anopheles midguts and sequenced both mosquito and parasite transcriptomes, profiling ~55,789 midgut cells and 3,495 parasites. Integrating new data with earlier scRNA-seq datasets produced a UMAP-based midgut atlas and a pseudotime trajectory capturing transitions from gametes/ookinetes to growing and segmenting oocysts. Gene-network clustering identified modules tied to cytoskeleton/motility, RNA processing and splicing, membrane/vesicle trafficking, translation/proteasome activity, aerobic respiration and apicoplast/lipid functions, and finally the inner-membrane complex and invasive organelle genes during sporozoite segmentation.
The authors used the expression maps to nominate druggable parasite factors. Delivering cipargamin or MMV670325 to infected mosquitoes via sugar from 2–10 dpi reduced oocyst size and, in many cases, eliminated salivary-gland sporozoites. A PfSIP2 conditional knockdown had no obvious effect on oocyst numbers or size but markedly reduced sporozoite invasion of human hepatocytes, indicating a role for an oocyst-expressed ApiAP2 in downstream infectivity. Dual-read single-cell barcodes and confocal microscopy revealed a consistent bias for parasites to be associated with progenitor ISC/EB cells at 36 hpi; later, confocal imaging showed visceral muscle fibres wrapping or stretching around large oocysts.
Overall, the dataset exposes understudied midgut-stage biology and supplies experimentally validated targets and testable hypotheses for transmission-blocking interventions and mosquito-targeted antimalarials.
Context and relevance
Transmission stages of P. falciparum are a major bottleneck in the parasite life cycle but are understudied because they are restricted to in vivo systems and occur at low density. This work fills critical gaps by profiling the parasite and vector simultaneously at single-cell resolution, linking parasite transcriptional programmes to midgut cell types and revealing interactions that may guide ookinete exit and oocyst establishment. The validated targets (PfATP4, PfLRS, PfSIP2) and pathways (membrane remodelling, respiration, translation) are directly relevant to development of transmission-blocking drugs, mosquito-based antimalarials and antigen/vaccine design aimed at blocking onward transmission. The finding that P. falciparum interacts with progenitor cells—and that oocysts associate with muscle—also reframes how we think about vector tolerance and parasite strategies to minimise damage to the mosquito host.
Author style
Punchy: this paper is dense with usable data and actionable results. If you work on malaria transmission, vector–parasite interactions, antimalarial target discovery or single-cell atlasing, the details and datasets here are worth digging into — the authors include validated drug interventions and a public single-cell resource to mine.
Why should I read this?
Short version: they mapped the parasite’s midgut life stages at single-cell resolution, found which genes drive the big transitions, validated drugs that stop oocyst growth and showed who the parasites hang out with in the mosquito. If you care about blocking transmission (and who doesn’t), this saves you time — the hard work is done and the datasets are public. Big win for people wanting new transmission-blocking targets or mosquito-targeted strategies.