Continuous cell-type diversification in mouse visual cortex development
Article metadata
Article Date: 05 November 2025
Article URL: https://www.nature.com/articles/s41586-025-09644-1
Article Image: 
Summary
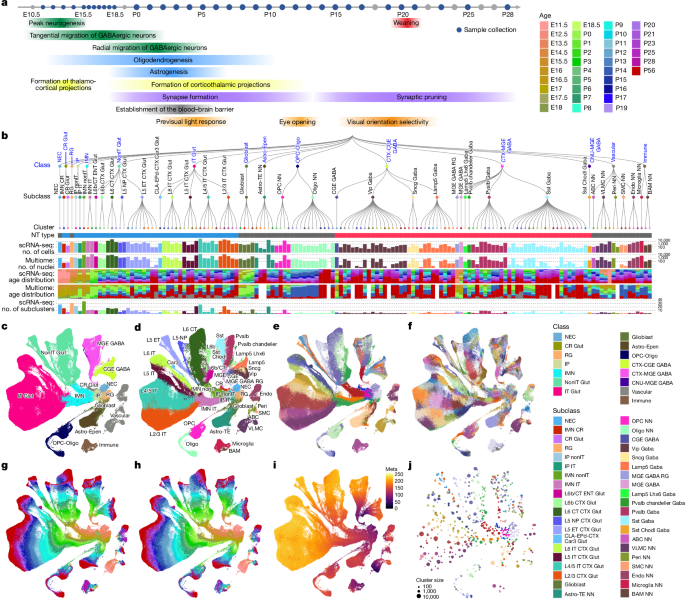
This Nature paper from the Allen Institute presents a dense, multimodal single-cell atlas of mouse visual cortex development from embryonic day 11.5 to adult (P56). Using ~913k single-cell transcriptomes and ~332k Multiome nuclei (paired snRNA‑seq + snATAC‑seq), the authors build a hierarchical taxonomy (15 classes, 40 subclasses, 148 clusters, 714 subclusters) and a time-resolved trajectory map showing continuous cell-type diversification.
Key findings include an early bifurcation between IT and nonIT glutamatergic lineages already visible at the intermediate progenitor/immature neuron stages, staggered but overlapping gliogenesis onset (glioblasts → astrocytes/OPCs around E15.5–E17), and prolonged postnatal diversification — with big bursts of new transcriptomic clusters after eye opening (P11–P15) and at the onset of the critical period (P20–P28).
The Multiome data link chromatin accessibility peak modules to developmental gene expression, enabling prediction of TF regulators and gene regulatory networks (GRNs). Temporal regulators (for example, Tcf4, Sox4, Mef2 family, AP‑1 complex) and cell-type TFs (Pou3f family, Nr4a2, Rora/Rorb, Sox family members) are highlighted as drivers that coordinate specification, maturation and activity-dependent refinement. The authors release processed datasets and supporting resources for further use.
Key Points
- A comprehensive single-cell and single-nucleus multimodal atlas of mouse visual cortex development (E11.5 → P56) with very high temporal resolution.
- Final taxonomy: 15 classes, 40 subclasses, 148 clusters and 714 subclusters across neuronal and non-neuronal types.
- Early, transcriptomic bifurcation between intratelencephalic (IT) and non‑IT glutamatergic fates appears at progenitor/IMN stages, not strictly by layer order.
- Glial trajectories (glioblast → astrocyte / OPC → oligodendrocyte) emerge concurrently with neuronal maturation; temporal TF programs for gliogenesis are identified (Rfx4, Rora/Rorb, Nr3c2, Sox family differences).
- Postnatal diversification is extensive: many adult-like clusters appear after birth, with bursts at eye opening and critical period — especially in IT excitatory neurons and Sst interneurons.
- Chromatin accessibility modules and peak–gene correlations link regulatory elements to target genes; SCENIC+-style GRN inference predicts activating and repressive TF–peak–gene triplets (e.g. AP‑1, Nr4a2, Pou3f family, Sox/Mef2 networks).
- Eye opening triggers widespread, cell-type-specific transcriptional and epigenomic changes including immediate‑early gene induction and shifts in synaptic/membrane-related programmes.
- All processed data and many resources are publicly available via BICAN/NeMO and the Allen-hosted links for reuse.
Context and Relevance
This work advances our understanding of how adult cortical cell types arise and diversify over time by combining ultra-dense temporal sampling with paired transcriptomic and chromatin data. It refines developmental models by emphasising early lineage bifurcation (IT vs nonIT), showing staggered but overlapping neurogenic and gliogenic programmes, and demonstrating a strong role for sensory-driven activity (eye opening) in shaping final cell-type diversification. The resource will be directly useful for researchers studying cortical development, circuit assembly, TF function, and for anyone mapping cell types across modalities or comparing across cortical areas and species.
Why should I read this?
Short version: if you care about how the cortex gets wired — and what makes a neuron become one specific type rather than another — this paper is gold. It's a massive, time-lapse atlas that tells you when cell identities appear, which genes and regulatory elements switch on, and which TFs likely drive those switches. It also shows that the biggest molecular changes happen after birth (hello, eye opening), so if you're into activity‑dependent development or TF networks, this saves you weeks of digging by giving the data and suggested regulators up front. Proper nerd-food for developmental neuroscientists.
Author style
Punchy — the study packs a lot of discovery and practical value: a high‑resolution map, predicted regulatory interactions and downloadable datasets. If you work on cortical cell identity, interneuron diversity, glial maturation or epigenetic regulation, read the full paper; the methods and supplementary tables are dense but hold actionable leads.
Source
Original article: Source: https://www.nature.com/articles/s41586-025-09644-1