Rare genetic variants confer a high risk of ADHD and implicate neuronal biology
Article Date: 12 November 2025
Article URL: https://www.nature.com/articles/s41586-025-09702-8
Article Image: Figure 1
Summary
This Nature study reports a large whole-exome sequencing analysis of ADHD that identifies rare, deleterious coding variants that substantially increase ADHD risk and point to neuronal mechanisms. The authors analyse 8,895 people with ADHD from the Danish iPSYCH cohort plus large control data (including 44,779 gnomAD exomes), find an excess burden of rare protein-truncating and severe missense variants in constrained genes, and identify three exome-wide significant genes: MAP1A, ANO8 and ANK2. Protein interaction, expression and single-cell analyses implicate synaptic biology and specific neuronal types (dopaminergic and GABAergic). Rare variants also associate with poorer education, lower socioeconomic status and reduced IQ among people with ADHD, and they act additively with common-variant polygenic risk.
Key Points
- Large whole-exome study: 8,895 ADHD cases (iPSYCH) versus 53,780 controls (iPSYCH + gnomAD exomes).
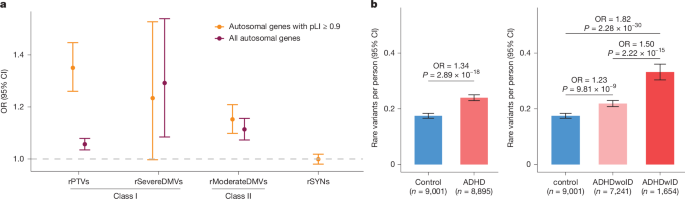
- Increased burden of rare protein-truncating variants (rPTVs) and severe damaging missense variants in constrained genes (pLI >= 0.9).
- Three exome-wide significant genes: MAP1A (OR ~13.3), ANO8 (OR ~15.3) and ANK2 (OR ~5.6); most top genes are evolutionarily constrained.
- PPI and IP-MS in neuronal models link the index proteins to synaptic, ribosomal and cytoskeletal processes and to networks enriched for autism and developmental-disorder risk genes.
- Single-cell analyses point to dopaminergic and GABAergic neurons (and medial neuroblasts) as relevant cell types for rare-variant risk.
- Rare deleterious variants correlate with worse life outcomes in ADHD: lower educational attainment, higher risk of low socioeconomic status and a drop in IQ per ultra-rare damaging variant.
- Joint analysis indicates rare and common variants act additively on ADHD risk; the rare-class I burden explains a small but measurable share of liability.
- There is substantial sharing of rare-variant risk genes between ADHD and autism, suggesting shared neurodevelopmental pathways.
Content summary
The authors sequenced exomes from almost 9,000 ADHD cases and used deep QC and comparison with 44,779 gnomAD non-psychiatric exomes to focus on ultra-rare, likely damaging variants (allele count <= 5). They defined two classes: class I (rPTVs + very damaging missense MPC>3) and class II (moderately damaging missense 2<=MPC<=3). The strongest enrichment was for class I variants in constrained genes (pLI >= 0.9). Gene-burden testing (after excluding genes with potential technical bias) identified MAP1A, ANO8 and ANK2 as exome-wide significant.
Protein interaction mapping in human iPS-derived neural progenitors and excitatory neurons revealed networks enriched for synaptic and cytoskeletal functions and for genes implicated in autism and developmental disorders. Expression analyses across BrainSpan and single-cell RNA-seq datasets showed high expression of top rare-variant genes across development and an enrichment of their expression in developing dopaminergic neurons and GABAergic neurons. Epidemiological linkage to Danish registers demonstrated associations between rare protein-truncating variants and lower education and SES among people with ADHD; a clinical sample showed a decrease in IQ per ultra-rare deleterious variant. Finally, combining ADHD polygenic scores with rare-variant status indicated additive effects on risk.
Context and relevance
This work goes beyond GWAS by pinpointing individual genes where rare coding lesions confer large effects. MAP1A suggests cytoskeletal and synaptic structure involvement; ANK2 and ANO8 implicate ion-channel or membrane transport processes, consistent with synaptic channelopathies. The convergence of rare and common signals on neuronal cell types, especially dopaminergic and GABAergic neurons, aligns with longstanding hypotheses about dopamine signaling in ADHD and with pharmacology (for example, methylphenidate’s action on dopamine transport).
Why should I read this?
Because if you want the short version: this Nature paper actually digs into the rare coding side of ADHD genetics and finds real genes that point at neurons, synapses and channels — not just statistical loci. It also links those rare hits to real-life outcomes like schooling, jobs and IQ. If you care about where ADHD biology might lead for mechanism studies or future targets, this is worth skimming (or reading properly if you like gene names and networks).
Author style
Punchy: big sample, clear signal, concrete genes. The study matters — it moves ADHD genetics from polygenic maps to specific high-impact coding variants and tangible biology. If you follow neurodevelopmental genetics or translational neurobiology, the details are worth digging into.