Viral NblA proteins negatively affect oceanic cyanobacterial photosynthesis
Article Date = 12 November 2025
Article URL = https://www.nature.com/articles/s41586-025-09656-x
Article Title = Viral NblA proteins negatively affect oceanic cyanobacterial photosynthesis
Article Image = https://media.springernature.com/lw685/springer-static/image/art%3A10.1038%2Fs41586-025-09656-x/MediaObjects/41586_2025_9656_Fig1_HTML.png
Summary
This study demonstrates that T7-like cyanophages commonly carry a small auxiliary gene, nblA, which they express early during infection to trigger targeted disassembly and degradation of the host phycobilisome antenna complex. Viral NblA expression reduces the photochemical quantum yield of PSII by roughly 50% during infection, accelerates phage replication (shorter latent period), and directs proteolytic cleavage across many phycobilisome subunits and other photosynthesis- and housekeeping-related proteins. The gene is widespread among oceanic T7-like cyanophages (especially clade B), and the authors estimate viral NblA may reduce picocyanobacterial light harvesting in the upper ocean by ~0.2–5% collectively.
Key Points
- Cyanophage-encoded nblA is transcribed and translated early during infection and differs in timing from the host nblA.
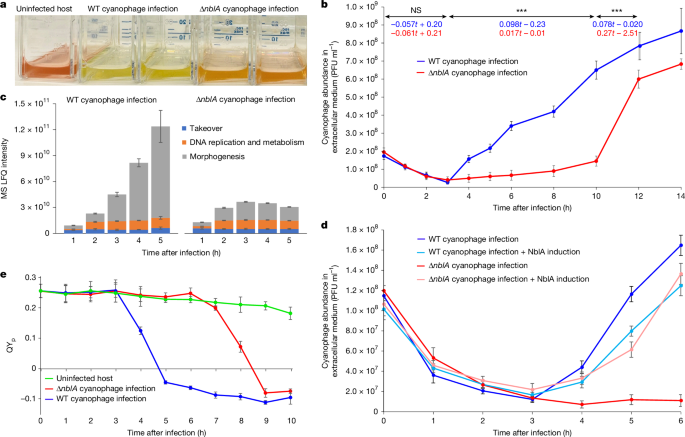
- Deletion of phage nblA (∆nblA) delays the phage latent period from ~3 h to 8–10 h and reduces production of morphogenesis proteins, showing nblA increases phage fitness.
- Wild-type phage infection causes rapid loss of energy transfer from phycobilisome peripheral disks to chlorophyll and a ~50% drop in PSII quantum yield; this effect is delayed in ∆nblA infections.
- Ectopic expression of viral NblA alone disrupts energy transfer and causes proteolytic cleavages in many phycobilisome subunits, confirming a direct role in antenna disassembly.
- N-terminomics reveals NblA-directed cleavages across almost all rod proteins, some core proteins and multiple photosystem and housekeeping proteins, suggesting broad proteolytic targeting (likely via Clp protease recruitment).
- About 46% of non-redundant T7-like cyanophage genomes encode nblA, concentrated in clade B; read recruitment shows 35–65% of T7-like cyanophages carry nblA depending on depth (surface vs DCM).
- Rough global estimates suggest these viral nblA genes could reduce picocyanobacterial light harvesting in the upper ocean by roughly 0.2–5%.
Content summary
Phycobilisomes are large, external light-harvesting antennae in many cyanobacteria that funnel light to photosystems. The small proteolysis adaptor NblA in cyanobacteria mediates controlled phycobilisome degradation under stress. The authors studied S-TIP37, a T7-like cyanophage infecting Synechococcus WH8109, and showed the phage nblA is strongly expressed from ~2 h post-infection, coincident with phage DNA metabolism genes.
Constructed ∆nblA cyanophage mutants show markedly reduced host bleaching and a much longer latent period; ectopic expression of the phage nblA in the host rescues the fast infection phenotype. Spectroscopy and fractionation demonstrate that WT phage infection rapidly disassembles phycobilisomes (intact complexes disappear) and interrupts energy transfer from peripheral PE disks to chlorophyll, while ∆nblA infections show delayed, partial disassembly.
N-terminal proteomics after induced expression of viral NblA identified >200 cleavage events enriched by NblA expression. Cleavages map across peripheral PEII and PEI disks, phycocyanin, linker proteins and even some core APC subunits, consistent with stepwise rod disassembly beginning at the exterior disks. Additional cleavages affect PSI and PSII-associated proteins and a range of translation, metabolic and housekeeping proteins, indicating broader host takeover beyond antenna degradation.
Genomic and metagenomic surveys find nblA common among T7-like cyanophages—particularly clade B—and also present in many picocyanobacterial genomes (including low-light Prochlorococcus). Read-recruitment to the Global Ocean Virome suggests a substantial fraction of abundant clade B phages carry nblA, especially at the deep chlorophyll maximum. Combining infection-level effects and environmental prevalence, the authors estimate a small but measurable global impact on picocyanobacterial light harvesting (up to ~5% in some layers).
Context and relevance
Why this matters: viral auxiliary metabolic genes (AMGs) are known to rewire host metabolism during infection; this work shows a small viral adaptor protein can directly dismantle the photosynthetic antenna and accelerate phage replication. That has implications for microbial ecology and biogeochemistry because cyanobacteria drive large portions of marine primary production. Viral targeting of the antenna frees amino acids for virion synthesis and can alter photosynthetic energy flow and dissolved organic matter composition after lysis. The finding that nblA is widespread in abundant cyanophage clades—more so at depth where cells invest heavily in antennae—suggests an ecologically relevant mechanism by which viruses modulate oceanic photosynthesis and carbon cycling.
Why should I read this?
Short version: it’s clever and it matters. These viruses carry a tiny tool that rips apart the host’s light‑harvesting kit, speeds up infection and likely shifts how much light cyanobacteria actually use across big swathes of the ocean. If you care about how viruses shape marine productivity, or how tiny genes can have outsized ecosystem effects, this paper saves you time — read it.