Prime editing-installed suppressor tRNAs for disease-agnostic genome editing
Article Date: 19 November 2025
Article URL: https://www.nature.com/articles/s41586-025-09732-2
Article Image: 
Summary
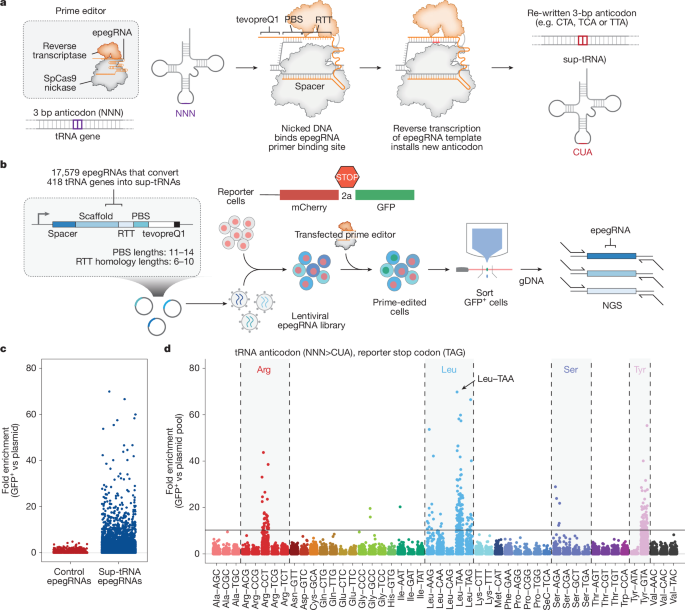
This study describes PERT (prime editing-mediated readthrough of premature termination codons), a one-time, disease-agnostic genome-editing strategy that converts endogenous human tRNA genes into suppressor tRNAs (sup-tRNAs) using prime editing. The authors screened and engineered variants across all 418 high-confidence human tRNA genes to identify and optimise sup-tRNAs that efficiently read through TAG (and in some cases TGA) premature stop codons when installed into the genome at native regulatory levels.
Through iterative optimisation (leader and terminator sequences, saturation mutagenesis of tRNA bodies and pegRNA architecture), they developed engineered Leu-TAA sup-tRNAs that, when installed at a single genomic copy, restore therapeutically relevant amounts of full-length protein or enzyme activity in multiple human cell disease models (Batten, Tay–Sachs, Niemann–Pick) and in vivo in mice (GFP reporter and Hurler syndrome model). PERT produced enzyme recovery sufficient to rescue pathology in the Hurler model, showed no detectable widespread off-target prime editing, and did not produce measurable proteome-wide readthrough of natural stop codons under tested conditions.
Key Points
- PERT uses prime editing to convert an endogenous tRNA gene into an optimised suppressor tRNA, avoiding exogenous high-level tRNA overexpression.
- Comprehensive screens (17,000+ epegRNAs per anticodon target plus leader/terminator and saturation-mutagenesis libraries) identified effective backbones—notably Leu, Arg, Tyr, Ser—for TAG or TGA readthrough.
- Engineered Leu-TAA sup-tRNAs achieved up to ~35% of wild-type protein yield from a single genomic copy in reporter assays and restored 20–70% enzyme activity in multiple disease-relevant cell models.
- PERT rescued disease phenotypes in vivo: co-delivered GFP reporter readthrough (~25%) and partial-to-therapeutic IDUA enzyme restoration (~6% in key tissues) produced near-complete rescue in a Hurler (Idua W392X) mouse model.
- Optimised prime editor designs (PE6 variants, PE3/PE3b strategies and MLH1 dominant-negative where appropriate) substantially improved installation efficiencies at endogenous loci.
- Extensive off-target and proteomic analyses found no significant genome-wide off-target prime edits and no detectable widespread readthrough of natural termination codons under the tested conditions.
- Pooled ClinVar-based reporter screening across 14,746 pathogenic TAG PTCs showed an average readthrough score of ~69%, indicating broad potential applicability for many nonsense mutations.
Context and relevance
Most current therapeutic genome-editing efforts are allele-specific and require bespoke reagents for each pathogenic mutation. PERT offers an allele-agnostic, potentially one-time genomic approach that could address classes of nonsense mutations across many genes with a single editing composition. This greatly reduces the need to develop thousands of separate therapies for nonsense-mutation diseases and fits current industry trends toward scalable, durable genome-editing treatments. The approach leverages the low expected perturbation of tRNA homeostasis when converting a single redundant tRNA gene and the high specificity of prime editing.
Why should I read this?
Want the short version: this paper shows a clever way to fix lots of different nonsense mutations with one edit. Instead of tailoring a fix for each broken gene, the authors turn one of your own tRNA genes into a stop-codon skipper — permanently. If you care about scalable genetic therapies, cystic fibrosis, muscular dystrophy, lysosomal storage disorders or the push to make genome editing more broadly applicable, this is exactly the kind of advance to skim now and read in detail later.
Author style
Punchy: the paper is a method-driven tour-de-force — deep, systematic screens, careful engineering and convincing in vivo proof-of-concept data. If you’re interested in translational genome editing, the results are significant: they demonstrate a realistic route to disease-agnostic rescue of many nonsense mutations and back it with safety and specificity assays that matter for clinical development.
Caveats and next steps
Key limitations include incomplete coverage for TAA stop codons, variable readthrough depending on local sequence context and the need to improve in vivo delivery and editing efficiencies for some tissues. Further work on delivery vehicles (capsid engineering, LNPs, VLPs), long-term safety and regulatory pathways is required before clinical translation.