Repulsions instruct synaptic partner matching in an olfactory circuit
Article Date: 19 November 2025
Article URL: https://www.nature.com/articles/s41586-025-09768-4
Main image: 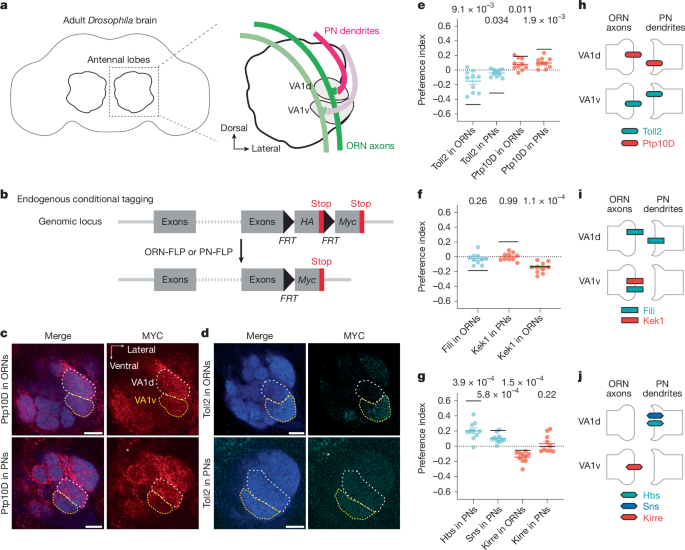
Summary
This study uses transcriptome-guided genetics in Drosophila to show that repulsive cell-surface protein (CSP) pairs actively prevent incorrect synaptic matches in the antennal lobe. The authors identify three CSP pairs with largely inverse expression in neighbouring glomeruli and show they act as trans-cellular repulsive cues that steer olfactory receptor neuron (ORN) axons and projection neuron (PN) dendrites away from non-partner partners. The pairs are Toll2–Ptp10D, Fili–Kek1 and Hbs/Sns–Kirre. Genetic knockdown or mutation of these molecules causes ORN/P N mistargeting; gain-of-function and suppression experiments support sender–receiver repulsion models. Only Hbs/Sns–Kirre showed clear direct binding in biochemical assays; Fili–Kek1 and Toll2–Ptp10D likely require cofactors or specific conditions. The authors argue that combinatorial, reused repulsive cues complement attractive cues to greatly improve wiring specificity across 50 ORN–PN pairs.
Author style: Punchy — this is a solid mechanistic advance on how repulsion, not just attraction, helps neurons pick the right partners.
Key Points
- Three CSP pairs with inverse expression across adjacent glomeruli (Toll2–Ptp10D, Fili–Kek1, Hbs/Sns–Kirre) act to prevent mismatching between non-partner ORNs and PNs.
- Loss-of-function (RNAi or mutants) in sender or receiver cell types causes ORN axons or PN dendrites to mistarget into neighbouring glomeruli.
- Gain-of-function (misexpression) produces repulsion phenotypes that are suppressed when the putative receiver is removed, supporting trans-cellular sender→receiver repulsion.
- Hbs/Sns–Kirre show direct heterophilic binding in vitro and tissue assays; Fili–Kek1 and Toll2–Ptp10D do not show direct binding in the authors’ assays, suggesting cofactors, post‑translational modifications or multimeric contexts may be required.
- Expression analysis shows these repulsive cues are reused combinatorially across many PN types, reducing mismatch errors while leaving room for partner attraction mechanisms.
- Mistargeted ORN axons are enriched for presynaptic markers, implying ectopic synapse formation when repulsive signals are perturbed.
- Approach combined single-cell transcriptomics, conditional endogenous tagging, in vivo genetic screens, sparse single-neuron manipulations and biochemical binding assays.
- Repulsion complements previously known attractive codes (e.g. teneurins, IgSF, cadherins) to sharpen synaptic partner choice during development.
Content summary
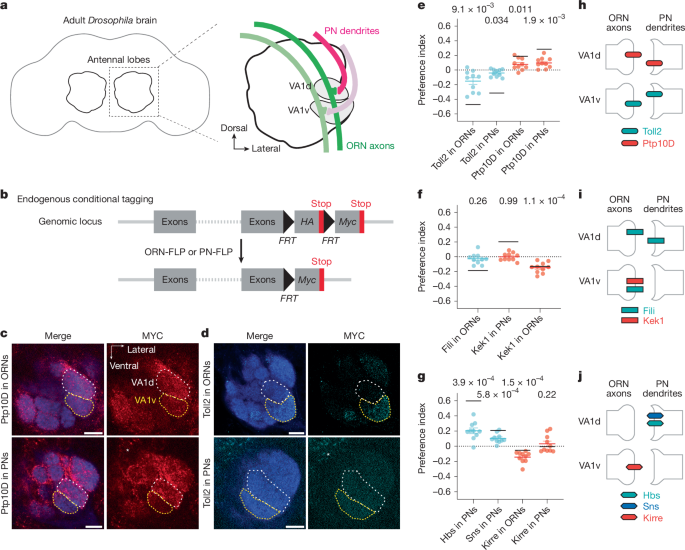
The authors started with single-cell transcriptomes at 24–30 h APF and a list of candidate CSPs enriched in the developing antennal lobe. A focused in vivo RNAi screen identified 14 candidates that perturb targeting; three CSP pairs stood out because they are expressed inversely between ORN and PN partners in neighbouring glomeruli VA1d and VA1v.
Using CRISPR knock-ins with conditional tags, the team mapped protein expression at 42–48 h APF and confirmed inverse patterns for the seven proteins across the two glomeruli. Cell-type-specific knockdowns and mutant analysis showed Ptp10D functions in VA1d-ORNs (and PNs) to prevent mistargeting into VA1v, while Toll2 acts in the neighbouring PN (VA1v) as a sender of repulsive signals. Similar sender–receiver roles were shown for Fili–Kek1 and Hbs/Sns–Kirre pairings affecting other ORN–PN pairings.
Functional assays (misexpression and suppression) support trans-cellular repulsive signalling: ectopic expression of a sender in the wrong cell repels partner neurites, and removing the receiver reduces that effect. Binding assays and tissue staining demonstrated direct Hbs/Sns–Kirre interactions, but Fili–Kek1 and Toll2–Ptp10D lacked detectable direct binding under the conditions tested, leaving biochemical mechanism open.
Finally, transcriptome and expression mapping indicate the three repulsive cues are used combinatorially across the antennal lobe, providing a compact, reusable code that reduces erroneous matches among 50 ORN–PN pairs. The authors propose repulsion plus attraction together improve both specificity and search efficiency during partner matching.
Context and relevance
Why it matters: neural circuits require extreme precision; until now most characterised synaptic matching cues were attractive. This work shows repulsion is a widespread, instructive mechanism for partner selection and is used combinatorially to scale specificity. For anyone studying neural development, synaptic specificity, or molecular logic of wiring, this paper provides both new molecular players and a conceptual shift — repulsive trans-cellular codes are integral, not peripheral, to partner matching.
Why should I read this?
Want to know how neurons avoid bogus hookups? This paper cuts the jargon and shows repulsive surface cues — not just attraction — actively steer axons and dendrites to the right partners. If you work on circuit assembly, adhesion/receptor biology or modelling wiring specificity, it’s worth a proper read: the experiments are clever, the genetic and single-cell strategy is neat, and the idea that repulsion is reused combinatorially is a tidy solution to a hard scaling problem.