The Microflora Danica atlas of Danish environmental microbiomes
Article Date: 03 December 2025
Article URL: https://www.nature.com/articles/s41586-025-09794-2
Article Image: https://media.springernature.com/lw685/springer-static/image/art%3A10.1038%2Fs41586-025-09794-2/MediaObjects/41586_2025_9794_Fig1_HTML.png
Summary
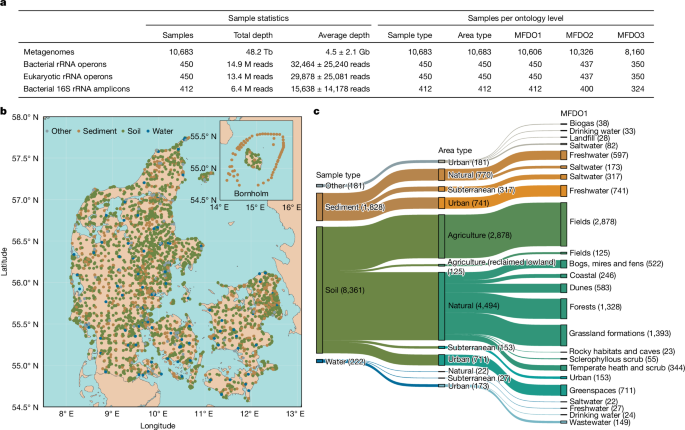
The Microflora Danica (MFD) project delivers a national atlas of environmental microbiomes across Denmark. The resource combines 10,683 metagenomic samples (Illumina shotgun; total ~48.2 Tb), long-read rRNA operon and UMI‑tagged nearly full-length 16S and 18S amplicons, and a purpose-built Microflora Global (MFG) 16S reference database. The sampling is geographically comprehensive, habitat‑classified with a five-level MFDO ontology, and includes GPS-linked, curated metadata.
Key outcomes: an estimate of terrestrial bacterial richness with a conservative lower bound of ~114,400 species (Hill richness), discovery of large species-level novelty (82.5% of bacterial OTUs novel vs SILVA at 98.7% clustering), recovery of 19,253 medium‑ or high‑quality MAGs (5,518 species-level clusters, ~4,604 novel vs GTDB), and identification/description proposals for two abundant uncultured nitrifier taxa: “Candidatus Nitrososappho danica” (an AOA linked to disturbed soils) and “Candidatus Nitronatura plena” (a widespread comammox Nitrospira in natural soils).
The MFG 16S database (≈30.2M sequences, clustered to 342,673 species-level OTUs at 98.7%) substantially improves read classification (genus-level classification of metagenomic 16S fragments: 46.1% with MFG vs 32.2% with GreenGenes2). The atlas also shows important ecological patterns: disturbed habitats (fields, greenspaces) can have high alpha diversity but low gamma diversity (community homogenisation), while natural habitats show greater gamma diversity and habitat breadth. Random-forest models using prokaryotic genus profiles can classify broad habitat types (MFDO1) reliably (agricultural soils PR‑AUC ~0.95), though fine-scale habitat classes remain challenging.
Key Points
- MFD is a national-scale microbiome atlas with 10,683 metagenomes, long-read rRNA operons and UMI 16S data, fully geo-referenced and habitat-annotated.
- The Microflora Global (MFG) 16S reference database (built from MFD plus public sources) improves taxonomic classification across datasets and habitats.
- Species-level novelty is high: ~82.5% of bacterial 16S OTUs are <98.7% identical to SILVA entries; eukaryotic 18S sequences likewise show ~77% novelty vs PR2.
- Conservative estimate of Danish terrestrial bacterial richness: ≥114,400 species (Hill richness) — a national baseline for monitoring.
- Recovered 19,253 medium/high-quality MAGs representing 5,518 species clusters; ~4,604 are novel compared with GTDB R220.
- Identified and proposed names for two ecologically important, previously undescribed nitrifiers linked to land use: “Candidatus Nitrososappho danica” (AOA) and “Candidatus Nitronatura plena” (comammox).
- Disturbed habitats show homogenisation (low gamma diversity) despite sometimes high local (alpha) richness — important for land-management and resilience considerations.
- Microbiome profiles can be used to classify broad habitat types and potentially monitor land-use or restoration changes, though higher-resolution classes are less distinct.
- All primary data, MAGs and reference resources are publicly available (NCBI BioProject PRJNA1071982, ENA long reads, Zenodo and GitHub links), enabling reuse.
Context and relevance
This study sets a new standard for national microbiome mapping: dense sampling across habitats, integration of long- and short-read sequencing, creation of an ecosystem-specific reference database, and recovery of thousands of genomes. It provides a baseline to track microbiome shifts from land management and climate change, informs biodiversity metrics by emphasising gamma diversity, and identifies candidate taxa with direct agricultural and greenhouse-gas implications (nitrifier groups linked to N2O emissions and fertiliser fate).
Author style
Punchy: this is big, systematic, and actionable. The paper doesn’t just catalogue microbes — it builds tools (MFG database, MAG catalogue, MFDO ontology) that make national‑scale microbiome monitoring and applied research (agriculture, restoration, greenhouse‑gas mitigation) much more tractable. If you work in environmental genomics, soil science, or national biodiversity monitoring, dig into the methods and datasets.
Why should I read this?
Short answer: because they did the hard yards so you don’t have to. If you want a validated national baseline of microbes, a better 16S reference for temperate habitats, genome resources for key functional groups (especially nitrifiers), or a dataset to test habitat‑classification and monitoring ideas — this is the one-stop shop. It’s a practical dataset with immediate uses in land‑management, climate and biodiversity research.