Gut micro-organisms associated with health, nutrition and dietary interventions
Summary
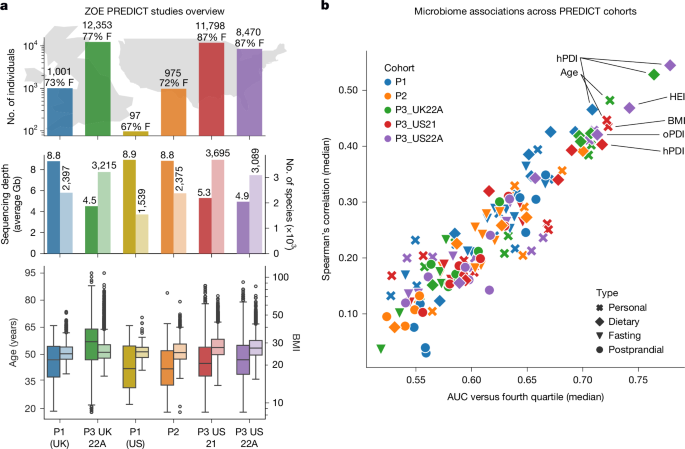
This Nature study analysed gut metagenomes and dietary/health data from five ZOE PREDICT cohorts (total n = 34,694 participants across the UK and USA) to map species-level associations between the microbiome, diet quality and cardiometabolic markers. The authors generated two public species rankings — the ZOE MB health-rank and the ZOE MB diet-rank — across 661 prevalent species-level genome bins (SGBs). Favourable and unfavourable species sets were identified, many favourable taxa are still uncharacterised, and the rankings correlate with BMI, disease status and respond predictably to dietary interventions (BIOME and METHOD trials).
Key Points
- Massive cross-sectional dataset: five harmonised ZOE PREDICT cohorts plus PREDICT1, totalling 34,694 participants with metagenomes, dietary indices and cardiometabolic markers.
- Two reproducible species-level rankings were produced: ZOE MB health-rank (based on anthropometric, fasting and postprandial markers) and ZOE MB diet-rank (based on validated diet indices).
- 661 prevalent SGBs were ranked; many top “favourable” species are uncharacterised (MAG-only), highlighting knowledge gaps.
- Rankings correlate with BMI and stratify healthy vs obese groups across external cohorts; healthy-weight individuals harbour more favourable SGBs.
- Meta-analyses across public disease cohorts show controls carry more favourable SGBs and fewer unfavourable SGBs (strongest signal for type 2 diabetes).
- Dietary interventions (prebiotic blend and personalised diet) produced predictable shifts: increased abundance of favourably ranked, fibre- and butyrate-associated species and decreases in unfavourable species.
- Rankings and code/data are publicly available (ZOE site, ENA and Zenodo) to support reproducibility and further research.
Content summary
The authors used partial Spearman correlations (adjusted for age, sex, BMI) and machine learning (random forest) to quantify consistent microbiome associations with 37 host markers grouped into personal, fasting and postprandial categories. They averaged cohort-level ranks to create the ZOE MB health-rank and derived a diet-rank from multiple validated diet indices. Top favourable SGBs were enriched for under-explored taxa (many MAG-only), while many unfavourable SGBs are known, cultured species previously linked to adverse outcomes. The rankings reproduce across countries for health associations, though diet associations show more country-specific heterogeneity. External validation across public cohorts confirmed associations with BMI and disease; dietary trials demonstrated that improving diet quality reproducibly shifts the microbiome toward more favourable-ranked species.
Context and relevance
This work addresses a core challenge in microbiome research: defining which species characterise a health-associated gut ecosystem in large, multi-centre Western cohorts. By linking long- and short-term dietary information with detailed cardiometabolic markers at scale, the study provides actionable species-level rankings that can be used to evaluate microbiome health signatures, design targeted dietary interventions and prioritise taxa for mechanistic follow-up. It is especially relevant to researchers in nutrition, microbiome therapeutics, and precision-health developers because the rankings are reproducible, validated longitudinally in intervention trials and publicly released.
Author note
Punchy take: this is a heavyweight, carefully controlled piece of work that turns huge metagenomic and diet datasets into a practical resource — the ZOE MB rankings — which you can use right away to assess or compare microbiome samples against diet- and health-linked species signatures.
Why should I read this?
Quick and informal: if you care about how diet ties into gut microbes and cardiometabolic risk, this paper is gold. It saves you the slog of combing small, inconsistent microbiome studies by providing a large, harmonised ranking of species tied to real dietary scores and health markers — and it shows those ranks actually move when diets improve. Useful whether you build interventions, design trials, or just want better biomarkers for gut health.