Mapping the genetic landscape across 14 psychiatric disorders
Summary
This Nature study from the Psychiatric Genomics Consortium maps shared and distinct common-variant genetic influences across 14 psychiatric disorders using large GWAS datasets and multiple complementary methods. The authors identify five stable genomic factors, numerous pleiotropic hotspots (notably a chr11 NCAM1–TTC12–ANKK1–DRD2 cluster), hundreds of factor-associated loci and many loci that differentiate one disorder from another. Functional follow-up implicates early neurodevelopment and specific brain cell types.
Key Points
- Analysed 14 psychiatric disorders using updated, larger GWAS datasets (mainly European-like ancestry).
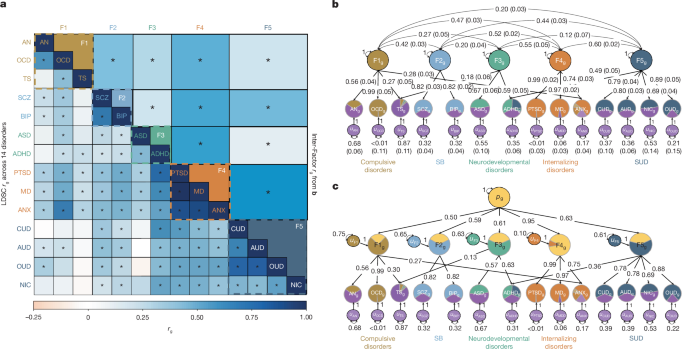
- Genomic structural equation modelling (Genomic SEM) recovered five latent factors: Compulsive, Schizophrenia/Bipolar (SB), Neurodevelopmental, Internalizing and Substance Use Disorders (SUD).
- A higher-order general psychopathology (_p_) factor was supported but did not fully capture cross-disorder heterogeneity.
- MiXeR showed pervasive polygenic overlap, mostly concordant in direction, beyond genome-wide genetic correlations.
- LAVA local analyses found 101 regional ‘hotspots’ of shared genetic signal; the top hotspot on chromosome 11 overlaps the NCAM1–TTC12–ANKK1–DRD2 cluster.
- Multivariate GWAS yielded 238 unique loci across the five factors and 160 hits for the _p_-factor; 48 factor loci were novel relative to univariate GWAS.
- Case–case GWAS (CC-GWAS) found 412 loci that distinguish pairs of disorders (109 LD-independent loci), most involving schizophrenia comparisons.
- Functional annotation points to pleiotropic variants acting in early neurodevelopment, with factor-specific enrichment in excitatory neurons, interneurons and glial cell types.
- Stratified genomic SEM highlighted enrichment in fetal brain marks, PI genes and specific neuronal subtypes for certain factors, supporting biological specificity.
- Primary limitation: analyses are dominated by EUR-like ancestry data; cross-ancestry conclusions are tentative pending larger diverse datasets.
Content Summary
The study integrates GWAS summary statistics for 14 DSM/ICD-defined psychiatric disorders, expanding prior cross-disorder efforts by adding six disorders (including three substance-use traits) and substantially increasing sample sizes. Using LDSC, MiXeR, Genomic SEM, LAVA, multivariate GWAS and CC-GWAS, the authors triangulate evidence of widespread genetic sharing alongside loci that uniquely differentiate disorders.
Genomic SEM produced a robust five-factor solution that explained the majority of common-variant genetic variance for many disorders and revealed strong interfactor correlations (notably Internalizing with SUD). A hierarchical _p_-factor captured broad transdiagnostic signal (particularly Internalizing traits) but exhibited greater SNP heterogeneity, indicating more nuanced subdomain structure is important.
Local (regional) analyses uncovered 101 pleiotropic hotspots; the most pleiotropic region on chr11 links to genes long-associated with behaviour and substance use. Multivariate locus discovery identified many factor-associated loci (including novel ones), while CC-GWAS revealed variants that differ between cases of distinct disorders — supporting genetic differentiation aligned with the five-factor model.
Functional follow-up (eQTL, Hi-C, single-cell enrichment, MAGMA, EWCE) points to early neurodevelopmental processes for broadly pleiotropic variants and to specific cell types — for example, excitatory neuron signatures for the SB factor and oligodendrocyte/glial signals for Internalizing traits — providing biological context for the factor structure.
Context and relevance
This paper advances cross-disorder psychiatric genomics by combining much larger samples, additional disorders and multiple analytic frameworks to move beyond simple pairwise genetic correlations. The five-factor structure is consistent with prior work but is now supported at greater scale and with finer, locus-level detail. Results bear on debates about diagnostic boundaries (for example, bipolar disorder versus schizophrenia), on shared pathophysiology across disorders, and on opportunities for drug repurposing that target transdiagnostic mechanisms.
For researchers, clinicians and pharma, the study provides a roadmap: pleiotropic, developmentally active loci and cell-type-specific signals are promising starting points for mechanistic studies and therapeutic exploration. For geneticists, the chr11 hotspot and the hundreds of factor loci are rich targets for follow-up.
Author style
Punchy: big, rigorous, and consequential — this is a field-defining cross-disorder genomic atlas that both confirms long-suspected overlaps and pinpoints where disorders diverge genetically. If you care about psychiatric nosology, neurobiology or translational targets, the details matter.
Why should I read this?
Short answer: because these folks did the heavy lifting so you don’t have to. If you want to know which psychiatric conditions share genetic roots, which genomic regions are hotbeds of cross-disorder risk, and where biology points (early brain development, specific neuron types), this gives you the map — and plenty of new loci to explore.