Nutrient requirements of organ-specific metastasis in breast cancer
Article Meta data
Article Date: 07 January 2026
Article URL: https://www.nature.com/articles/s41586-025-09898-9
Article Title: Nutrient requirements of organ-specific metastasis in breast cancer
Article Image: Figure 1
Summary
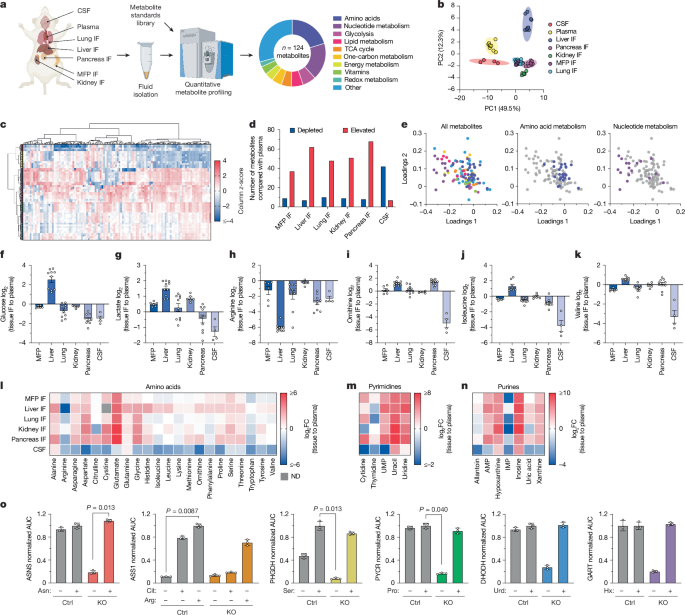
This Nature study maps microenvironmental nutrient landscapes and tests whether single nutrient levels determine where triple-negative breast cancer (TNBC) cells can form metastases. The authors quantified 124 metabolites in plasma, CSF and interstitial fluid from multiple mouse tissues, created TNBC cell lines engineered to be auxotrophic for specific metabolites (asparagine, arginine, serine, proline and nucleotides), and measured metastatic growth after intracardiac injection or direct implantation (brain and mammary fat pad).
Key results: tissue metabolite levels vary strongly by organ; single-nutrient availability alone does not reliably predict metastatic site preference; de novo nucleotide synthesis (GART, DHODH) is broadly essential for tumour growth across sites; amino-acid dependencies are heterogeneous and cell-line specific; and combined metabolite signatures (not single nutrients) correlate with metastatic potential. Dietary depletion of serine/glycine changed metastasis in a site-specific manner, underscoring the complexity of nutrient–tumour interactions.
Key Points
- Comprehensive profiling: 124 metabolites quantified in plasma, CSF and tissue interstitial fluid (mammary fat pad, liver, lung, kidney, pancreas) in mouse models, revealing distinct tissue nutrient landscapes.
- Engineered auxotrophs: TNBC cells (MDA-MB-231, HCC1806, EO771) with CRISPR knockouts for ASNS, ASS1, PHGDH, PYCR1/2/3, DHODH and GART allowed direct tests of nutrient dependence in vivo.
- Single nutrients fail as predictors: levels of individual metabolites in isolation generally do not predict whether a corresponding auxotroph can colonise a tissue.
- Nucleotide synthesis is critical: GART and DHODH knockouts showed strong defects across many tissues, indicating a broad requirement for de novo purine/pyrimidine synthesis despite extracellular nucleotides being present.
- Amino-acid dependencies vary: asparagine, arginine, serine and proline requirements were heterogeneous between cell lines and tissues, showing cell-intrinsic wiring matters.
- Multiple-metabolite signatures matter: 23 metabolites (including serine, glycine, tyrosine, glutamate, inosine, thymidine) consistently correlated with metastatic potential across cell lines — implying a combinatorial influence of nutrients.
- Microenvironmental compensation: residual amino-acid concentrations, stromal support, transporter expression and macromolecule scavenging can help tumours grow in ostensibly nutrient-poor sites.
Content summary
The team first validated stable, reproducible LC–MS measurement of 124 metabolites in plasma, CSF and tissue interstitial fluid from NSG and C57BL/6 mice. PCA and clustering showed tissue IF differs from plasma and CSF; nucleotide species contributed strongly to separation. Brain CSF was comparatively low in many amino acids, whereas some metabolites were higher in tissue IF than plasma.
Using CRISPR, the authors made auxotrophs lacking enzymes for asparagine (ASNS), arginine (ASS1), serine synthesis (PHGDH), proline (PYCR1/2/3) and nucleotide synthesis (DHODH, GART). Auxotroph phenotypes were validated in vitro and retained after in vivo outgrowth. Intracardiac injection assays and direct brain/MFP implants showed that nucleotide-synthesis genes (GART, DHODH) are broadly essential; amino-acid requirements were variable and cell-line dependent. Correlation analyses found only a few direct links between a single metabolite level and auxotroph dependency.
13C-glucose tracing in MDA-MB-231 tumours showed higher labelling of several amino acids in brain tumours versus MFP, and different nucleotide synthesis patterns, yet these activity measures did not reliably predict pathway dependency. A serine/glycine-deficient diet lowered tissue serine/glycine but affected metastasis at some sites (bone, kidney, ovary) and not others (brain, liver, lung), again showing site-specific effects.
DepMap correlations suggested CRISPR dependency scores for DHODH and GART align with metastatic potential to some tissues (notably lung), whereas expression or single metabolite levels were poorer predictors. Overall, metastatic preference emerges from a combination of multiple nutrient levels plus cell-intrinsic metabolic wiring and adaptive mechanisms such as salvage, transporter upregulation and stromal support.
Context and relevance
This work challenges the simple model that low availability of a single metabolite in a tissue dictates metastatic colonisation. Instead, it demonstrates that (1) tumour metabolic vulnerabilities are shaped by both the multi-metabolite composition of the microenvironment and intrinsic cell programmes, and (2) targeting metabolism clinically will require integrating nutrient availability, salvage pathways, stromal interactions and genetic dependencies. The clear, systematic mapping of tissue metabolites and in vivo functional testing make this high-value for researchers working on cancer metabolism, metastasis biology and metabolic therapy strategies.
Author style
Punchy — the authors did the heavy lifting: quantitative metabolomics across tissues plus functional CRISPR auxotroph tests in vivo. If you care about whether measuring one metabolite can reveal a therapeutic target for metastasis, this paper shows it rarely can; you need the bigger metabolic picture.
Why should I read this
Short version: want to know whether a low level of ‘X’ in the brain or liver means your drug targeting X will stop metastasis? Probably not. This paper saves you time by showing that single-nutrient snapshots rarely predict metastatic success — and points you to where to look instead (nucleotide synthesis, combined metabolite signatures, cell-intrinsic wiring and salvage mechanisms).
Source
Source: Nutrient requirements of organ-specific metastasis in breast cancer — Nature
Data & availability
Metabolomics datasets deposited to MetaboLights (MTBLS13151, MTBLS13152, MTBLS13164, MTBLS13165). Source data and supplementary material available at the article link.