Isolation, engineering and ecology of temperate phages from the human gut
Summary
This Nature study used a culture collection of 252 human gut bacterial isolates and multiple induction conditions to identify and validate inducible temperate phages. The team combined pure-culture inductions, synthetic community co-cultures and co-culture with human colonic epithelial (Caco2) cells, sequencing viral-enriched fractions to recover 125 inducible prophages representing 63 phage species. Key discoveries include that only a minority of predicted prophages are readily inducible in standard lab conditions, human cell products can trigger prophage induction, diversity-generating retroelements (DGRs) are common in Bacteroidota prophages and genetic inactivation of excision/integration genes can permanently trap prophages. The authors also describe a putative Hankyvirus genus and experimentally validate cross-host activity for several phages.
Key Points
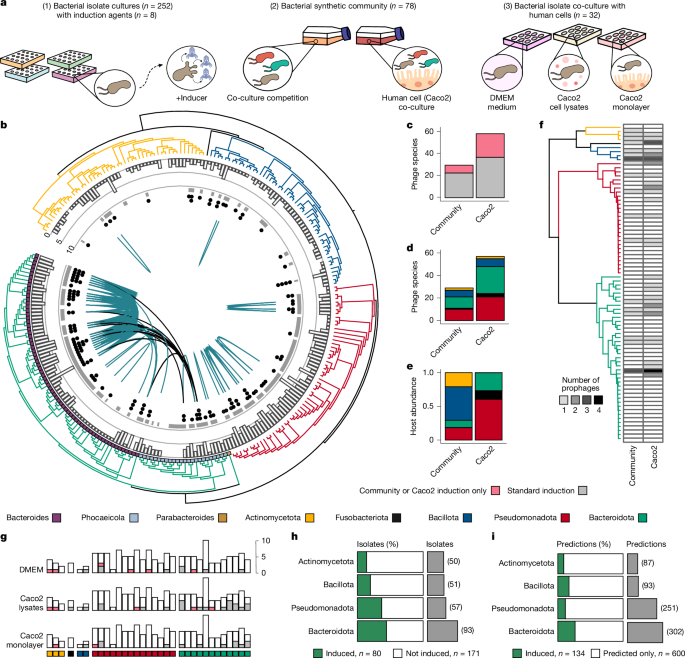
- Large-scale culture-based screen: 252 gut bacterial isolates across five phyla were tested under ten induction conditions, producing 125 inducible prophages from 73 isolates.
- Only a fraction of predicted prophages are inducible: ~18% of high-quality prophage predictions were induced in pure cultures; synthetic community and human-cell co-culture raised detectable induction but many prophages remained dormant.
- Human cellular products induce prophages: Caco2 lysates and monolayers revealed induction not seen with standard chemical inducers, suggesting host-derived factors matter.
- Diversity-generating retroelements (DGRs) are frequent, especially in Bacteroidota prophages, and appear linked to host-range expansion via tail-protein diversification.
- Genetic inactivation traps prophages: elevated mutation rates and non-synonymous changes in integrase/excision genes were found in non-induced prophages; deletion of a transposition gene in ΦPomma abolished induction experimentally.
- Polylysogeny matters: lysogens with multiple prophages (common in Bacteroidota) showed higher propensity for induction across conditions and may promote synchronous induction dynamics.
- Taxonomy and prevalence: most induced phages are Caudoviricetes; several species (including LoVEphage and Hankyvirus members) are detectable across human gut viromes.
Content summary
Researchers combined computational prediction with extensive experimental induction. They exposed isolates to mitomycin C, hydrogen peroxide, Stevia, carbon and SCFA starvation, and to human cell environments. From viral-enriched sequencing they confirmed which prophage regions entered lytic replication and assembled temperate phage genomes. Taxonomy was assigned using gene-voting and gene-sharing networks; about a quarter of phages could be placed in ICTV-accepted families. The study highlights differences between high-quality prophage predictions and those that are functionally inducible: induced prophages are enriched for structural and lysis genes, whereas many predicted elements look like remnants or accessory-rich cryptic regions. Through comparative genomics they link increased dN/dS in integration/excision genes to prophage inactivation, and they validate causality by CRISPR-based deletion of a transposition protein in ΦPomma, which prevents induction by Stevia and hydrogen peroxide.
Context and relevance
Temperate phages shape gut microbial ecology via lysogeny, lysis and horizontal gene transfer. While metagenomics has catalogued huge virome diversity, culture-based validation of active temperate phages has been limited. This work connects predicted prophage sequences to active induction events, shows that host (human) factors can trigger prophage activation, and provides a validated phage–host collection suitable for synthetic biology and microbiome research. The findings have implications for understanding microbiome stability, inflammation-associated virome shifts, and the potential to engineer phage dynamics or resistances in therapeutic contexts.
Why should I read this?
Short version: if you care about how phages actually behave in the gut (not just what sequences show up in metagenomes), this paper saves you time — it’s a wet‑lab, sequence‑verified tour of which prophages can be woken up, how human cells can flick the switch, and why many prophages are effectively trapped. Great if you work on microbiome ecology, phage therapy or phage engineering.
Author style
Punchy and consequential: the paper experimentally pins down induction triggers and genetic routes to prophage domestication, providing both ecological insight and engineered‑phage tools. If you work in the field, the validated phage–host pairs are worth a close look.