NIST Releases Trove of Genetic Data to Spur Cancer Research
Summary
NIST has released a comprehensive, multi-terabyte genomic dataset from a pancreatic cancer cell line whose donor explicitly consented to public sharing. The dataset includes results from 13 cutting-edge whole-genome sequencing technologies and matched non-cancerous cells, enabling quality control, method comparison, AI training and broader cancer research while avoiding past consent controversies.
Key Points
- The dataset is several terabytes and derived from a pancreatic tumour donated with explicit public-consent by a 61-year-old patient.
- NIST used 13 distinct whole-genome measurement technologies and published separate results for each method to aid comparison and validation.
- The resource helps clinical labs perform quality control and verify sequencing equipment and pipelines against a robust reference.
- Researchers can train AI models on the data to detect cancer-associated mutations and predict therapeutic responses.
- The matched normal (non-cancerous) cells from the same donor were also released, enabling tumour-normal comparisons.
- NIST’s open release avoids ethical issues tied to historic cell lines harvested without consent (for example, Henrietta Lacks) and sets a consented standard.
Content Summary
NIST’s Genome in a Bottle Consortium, through its Cancer Genome in a Bottle programme, published an extensively sequenced pancreatic cancer cell line and described it in Scientific Data. The dataset contains raw and processed outputs from each sequencing technology used, allowing users to cross-check their own sequencing results and identify discrepancies. The availability of a matched normal sample enhances the dataset’s value for variant calling and downstream analyses.
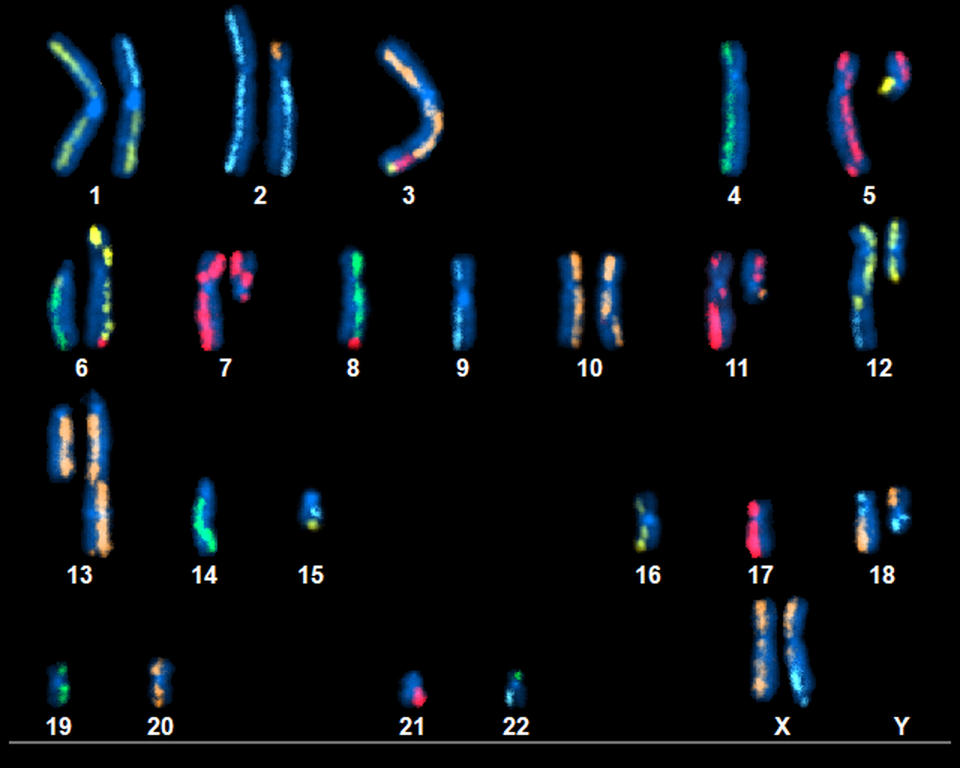
The release aims to support multiple use cases: quality assurance for clinical sequencing labs; benchmarking and improvement of existing sequencing platforms; training and validation datasets for machine-learning models that predict pathogenic mutations or drug sensitivities; and broader biological investigations into cancer development and chromosomal rearrangements, as illustrated by karyotype images showing chromosomal swaps in the tumour.
Context and Relevance
This is an important step for genomic standards in oncology. By publishing a consented, well-characterised tumour-normal pair sequenced with many modern techniques, NIST provides a reproducible reference that addresses both technical and ethical gaps in prior public resources. The dataset feeds into ongoing trends: rising regulatory focus on clinical sequencing accuracy, rapid adoption of AI in genomics, and increased demand for consented, shareable biomedical data.
Manufacturers can use the data to spot method-specific weaknesses and innovate; clinical labs can improve confidence in patient results; researchers can probe mutation patterns and structural rearrangements; and data scientists get a high-quality training resource for models that could speed up diagnostics and therapy selection.
Why should I read this?
Short version: if you work with genomic data, cancer diagnostics, sequencing tech or AI for biology, this is gold. NIST has done the heavy lifting—sequenced a consented tumour with lots of methods and made everything public—so you don’t have to reinvent the wheel. It’s a practical tool for checking your kit, training models, or exploring tumour biology without legal headaches.