Secretome translation shaped by lysosomes and lunapark-marked ER junctions
Article Meta
Article Date: 05 November 2025
Article URL: https://www.nature.com/articles/s41586-025-09718-0
Article Title: Secretome translation shaped by lysosomes and lunapark-marked ER junctions
Article Image: Figure 1
Summary
This Nature study shows that translation of secretory and membrane (secretome) mRNAs is not uniformly distributed across the endoplasmic reticulum (ER) but is concentrated at specific ER junctions marked by the ER-shaping protein lunapark (LNPK). Those LNPK-positive junctions frequently sit next to lysosomes and act as organisational hubs that boost ribosome occupancy and translation initiation on secretome mRNAs. Loss of LNPK selectively reduces membrane/secretory protein synthesis by impairing eIF2-dependent initiation; this defect is rescued by ISRIB, indicating involvement of an ISR-like pathway. Lysosomal activity and proximity further modulate this local translation, especially during amino acid starvation, suggesting lysosomes supply local resources that sustain co-translational translocation at junctions.
Key Points
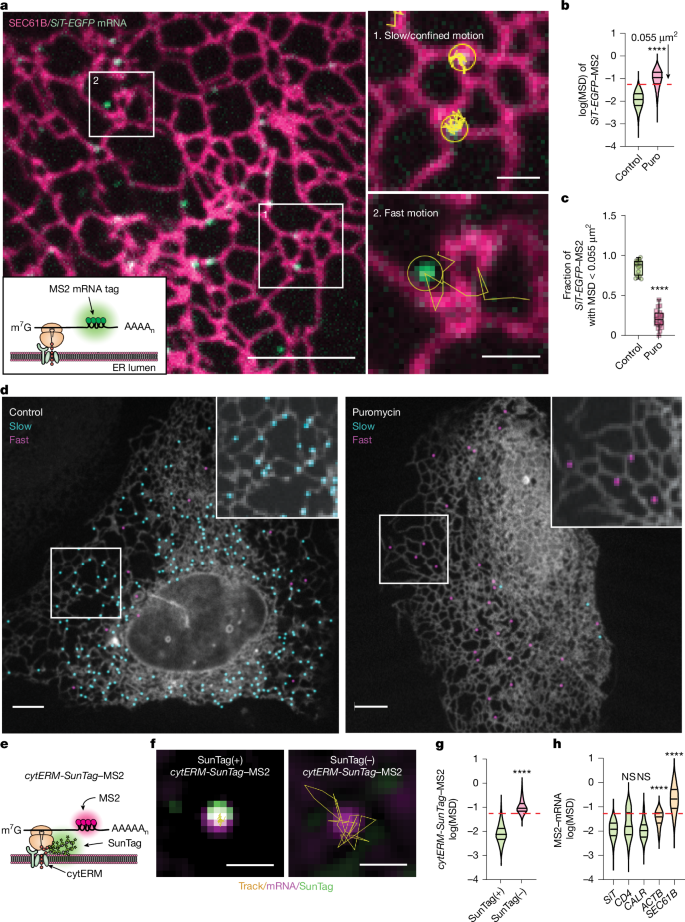
- Secretome mRNAs engage in confined, slow motion on the ER when actively translating; mobility increases when translation is disrupted (puromycin).
- Translating secretome mRNAs and slow-moving ribosomes are enriched at ER tubule junctions, not only on smooth ER sheets.
- LNPK marks a subset of ER junctions where translation is concentrated; translating mRNAs colocalise with LNPK signal.
- LNPK loss (KD/KO) reduces the translating fraction of secretome mRNAs and lowers membrane protein output while leaving global translation largely intact.
- LNPK-enriched junctions preferentially recruit lysosomes; lysosome proximity increases ribosome occupancy on nearby secretome mRNAs.
- Lysosomal proteolysis and acidic pH are required for the local translation boost; amino acid starvation intensifies lysosome-dependent localisation of translation.
- The translational enhancement depends on canonical initiation (sensitive to eIF2 regulation) — CrPV IRES reporters that bypass initiation control are unaffected by LNPK loss.
- ISRIB restores translation in LNPK-deficient cells, linking the phenotype to eIF2-dependent initiation and an ISR-like signal distinct from full canonical ISR activation.
Context and relevance
This work revises the textbook picture that secretory protein translation predominately occurs on perinuclear rough ER sheets. Instead, it identifies peripheral ER junctions — architecturally stabilised by LNPK and functionally coupled to lysosomes — as spatially defined hotspots for secretome synthesis. The paper bridges ER morphogenesis, inter-organelle contact biology and translational control, and it is relevant to researchers studying protein biogenesis, nutrient sensing, organelle contact sites, and neuronal or immune-cell secretory demands. The methods (single-molecule MS2/SunTag reporters, ribosome tracking, optogenetic ER–lysosome recruitment, proteomics and RNAseq) provide a robust multi-modal demonstration that local environment and organelle signalling shape translation efficiency.
Why should I read this?
Short answer: because it flips the simple idea of where secretory proteins are made. If you care about how cells tune secretory output — whether for basic cell biology, neurobiology or immune differentiation — this paper gives you the who (LNPK), where (ER junctions), and how (lysosome-linked, eIF2-dependent initiation). Read it to see the live-imaging tricks and the rescue with ISRIB — neat experiments that actually link structure, organelle cross-talk and translation control. We skimmed the figures and pulled the essentials so you don’t have to.
Author style
Punchy: the authors present a tight, mechanistic story with clear functional consequences — LNPK-marked ER junctions and nearby lysosomes form a localised platform that tunes secretome translation via initiation control. This is significant: it explains how cells spatially prioritise membrane/secretory protein synthesis and links organelle contacts to nutrient-responsive translational regulation. If your work touches translation, ER organisation, lysosome biology or stress responses, the detailed experiments here are worth digging into.