Lineage-resolved atlas of the developing human cortex
Article Date: 05 November 2025
Article URL: https://www.nature.com/articles/s41586-025-09033-8
Article Image:
Summary
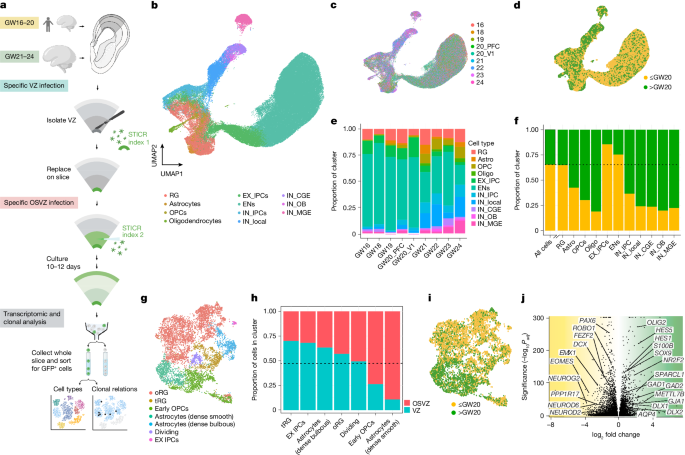
This study builds a lineage-resolved single-cell atlas of the human developing neocortex across midgestation using STICR, a high-throughput clonal barcoding method combined with scRNA-seq. The authors traced 6,402 neural stem and progenitor cells (and profiled ~97,500 single cells passing QC) from nine specimens spanning before and after midgestation (around GW20). They recover lineage barcodes in ~60% of cells and map clonal relationships to reveal how progenitor outputs change with developmental age and germinal niche.
Three major discoveries emerge: a midgestation switch from predominantly glutamatergic to locally generated GABAergic neurogenesis; truncated radial glia (tRG) are multipotent and importantly contribute to glutamatergic neurogenesis by generating intermediate progenitor cells (IPCs); and late-stage VZ/ISVZ progenitors still produce glutamatergic neurons with transcriptomic similarity to deep-layer/subplate neurons, implying a late phase of deep-layer-like neurogenesis.
Key Points
- Massively parallel lineage tracing (STICR) + scRNA-seq on mid-gestation human cortical slices yields a high-resolution clonal atlas (~97,540 cells; ~60% barcode recovery).
- A developmental shift around midgestation (after GW20) shows increased locally produced GABAergic-lineage cells; many are born dorsally and form clones shared with excitatory lineages.
- Truncated radial glia (tRG), primarily VZ-derived, are multipotent: they self-renew and generate IPCs and excitatory neurons as well as glia and inhibitory-lineage cells.
- VZ/ISVZ progenitors remain neurogenic late in the second trimester; some late-born glutamatergic neurons express deep-layer/subplate markers (TBR1, CTGF, NURR1), suggesting continued subplate/deep-layer neurogenesis.
- Gliogenesis onset is gradual and overlaps with ongoing neurogenesis; dorsally derived oligodendrocyte precursor cells (OPCs) and glial lineages are produced alongside neurons.
- Findings highlight human-specific and protracted developmental programmes that differ from mouse, with implications for disease modelling and understanding cortical expansion.
Context and relevance
This work addresses key unknowns about human cortical development that matter for basic neuroscience and for models of neurodevelopmental disorders. By combining clonal barcoding with single-cell transcriptomics across developmental timepoints and germinal niches, the authors clarify which progenitors produce which cell types and when — including unexpected local production of inhibitory neurons and sustained production of deep-layer-like glutamatergic neurons late in the second trimester. That’s important because many genes linked to autism and other developmental disorders are expressed in midfetal cortical neurons; knowing the lineage and timing refines where and when perturbations might act.
The atlas also reinforces differences between mice and humans: prolonged neurogenesis, dorsal generation of some interneurons, and active tRG contributions — all of which are crucial when translating findings from model organisms to the human brain or designing human cellular models.
Author style
Punchy take: big data, bold conclusions. The paper reads like a carefully executed detective story — dense experimental work that forces us to rethink assumptions about when and where human cortical neurons (and glia) arise. If you work on cortical development, neurodevelopmental disorders, organoids or comparative neurobiology, delve into the figures and methods; there’s lots of actionable detail.
Why should I read this?
Short answer: because this paper saves you months of digging. It gives you a mapped set of who-begets-who in the human cortex around midgestation — and drops a couple of surprises (local interneuron production, neurogenic tRG, late deep-layer-like neurons). If you care about human-specific development, modelling disease, or interpreting single-cell maps, this is exactly the sort of dataset and analysis you want to know about.