Conservation and alteration of mammalian striatal interneurons
Summary
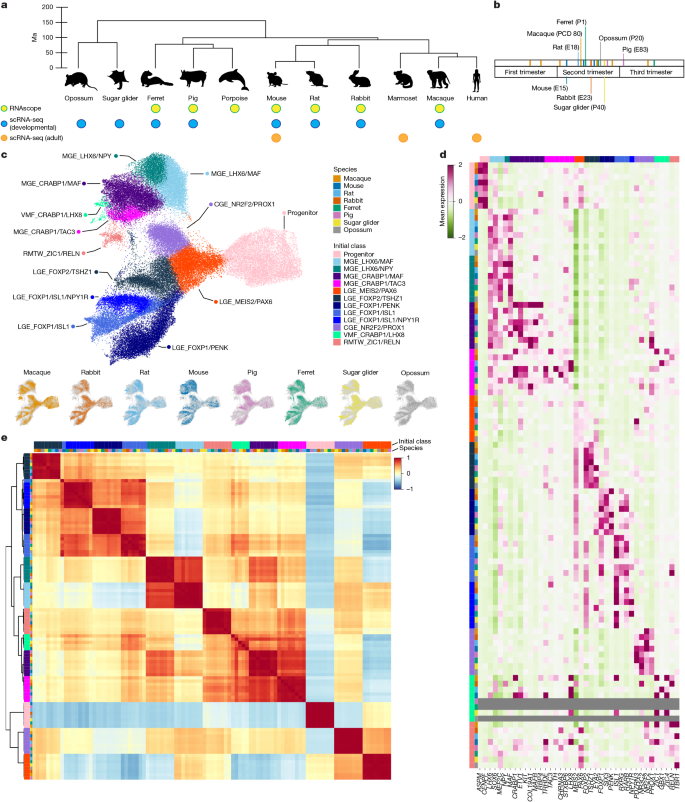
This study compares developing and adult inhibitory neurons across a broad set of mammals and shows that the major “initial classes” of striatal interneurons are conserved across placental mammals. A population named by TAC3 in primates is present broadly but has been modified in some clades — notably, rodents substitute TH expression (Th/Tac2 shifts) and show different distributional patterns. The team used high-coverage single-cell RNA sequencing across species, RNAscope in situ validation, cross-species integration (Harmony, scVI, SAMap) and module projection (Hotspot) to map homologies and lineage-specific changes in marker expression and migration.
Key Points
- Initial inhibitory neuron classes are conserved across placental mammals, segregating by cell type more than by species.
- The MGE_CRABP1/TAC3 initial class, previously thought primate-specific, is ancestral and found across Laurasiatheria and Marsupialia, but its allocation and markers vary by clade.
- Rodents show a derived modification: TAC3 (Tac2) expression is reduced and TH (Th) expression increased in the homologous lineage, producing Th-expressing interneurons in mouse and rat striatum.
- Pigs and ferrets show migration of MGE_CRABP1/TAC3 cells into cortex during development, whereas primates retain this population mainly in striatum.
- Adult primate TAC3 interneurons are homologous to mouse Th interneuron clusters, despite gene-expression shifts and differences in abundance and spatial distribution.
- Marker-gene turnover (including signalling-pathway genes like CRABP1, CER1) highlights evolutionary modifications on a conserved developmental scaffold rather than wholesale creation of new early cell types.
- Findings enable more confident translation of functional work between rodent Th interneurons and primate TAC3 interneurons, with caveats about marker differences and population abundance.
Content summary
The authors collected scRNA-seq data from developing cortex and striatum across a taxonomically broad set of mammals (macaque, pig, ferret, mouse, rat, rabbit, opossum, sugar glider) at stages corresponding to peak interneuron specification. They isolated inhibitory neurons, annotated MGE/LGE/CGE origins, and integrated datasets after downsampling to detect conserved “initial classes.” Integration and correlation analyses show strong conservation of initial classes across species and similar developmental trajectories.
Focusing on the MGE_CRABP1 family, they find two related initial classes: MGE_CRABP1/MAF and MGE_CRABP1/TAC3. The TAC3 class appears in many mammals but exhibits clade-specific marker turnover: in many rodents TAC3 (Tac2) is largely lost and replaced by Th expression early in development. RNAscope validated the presence and spatial patterns across species, demonstrating migration differences (pigs/ferrets send CRABP1/TAC3 cells into cortex) and persistence into adulthood in multiple taxa. Cross-species adult integrations and module projections confirm homology between primate TAC3 and mouse Th interneurons despite divergent gene markers and population sizes.
Context and relevance
This paper addresses a core question in comparative neurobiology: how do neuronal cell types evolve? The results support a model in which evolution acts on a conserved repertoire of early cellular classes and alters gene expression, numbers and migratory fate to generate species differences. For neuroscientists using rodent models, the study is especially relevant because it clarifies which rodent interneuron populations are homologous to primate cell types (for example, mouse Th interneurons as counterparts of primate TAC3 types), and flags marker-gene turnover that could mislead simple marker-based comparisons. It also suggests candidate conserved functional modules (acetylcholine receptors, TRH genes) that may underlie shared roles across species.
Why should I read this?
Short version: if you work with inhibitory neurons, evolution, or translational rodent models, this paper saves you time. It shows that the cell types you’re studying in mice often have direct counterparts in primates — but the usual marker genes can change. That means functional insights from rodents can be relevant, provided you heed gene-expression shifts and distribution differences.
Author style
Punchy: the authors cut through taxonomy noise with deep single-cell coverage and cross-species integration. They make a strong claim — mammalian brain evolution tweaks a shared developmental repertoire rather than inventing many brand-new early cell types — and back it with multiple validation methods. If you care about cell-type homology or translating rodent data to primates, this is important reading.