Spatial dynamics of brain development and neuroinflammation
Article Date: 05 November 2025
Source URL: https://www.nature.com/articles/s41586-025-09663-y
Article image: Figure 1
Summary
This study introduces and applies high-resolution spatial tri-omic technologies (DBiT-based spatial ARP-seq and spatial CTRP-seq) together with multiplex CODEX imaging to map chromatin accessibility or histone modification (H3K27me3), transcriptomes and ~150 proteins in the same tissue sections. The authors profiled mouse postnatal brains (P0–P21), benchmarked human V1 developmental samples, and examined focal demyelination/remyelination in an LPC mouse model. Key outcomes: chromatin accessibility can spread beyond RNA expression domains (epigenetic memory/priming), myelination programmes are spatially and temporally primed before protein appearance, corpus callosum (CC) myelination shows lateral-to-medial and bidirectional features, and focal lesions elicit both local and delayed distal microglial activation along white-matter tracts with distinct microglial states.
Key Points
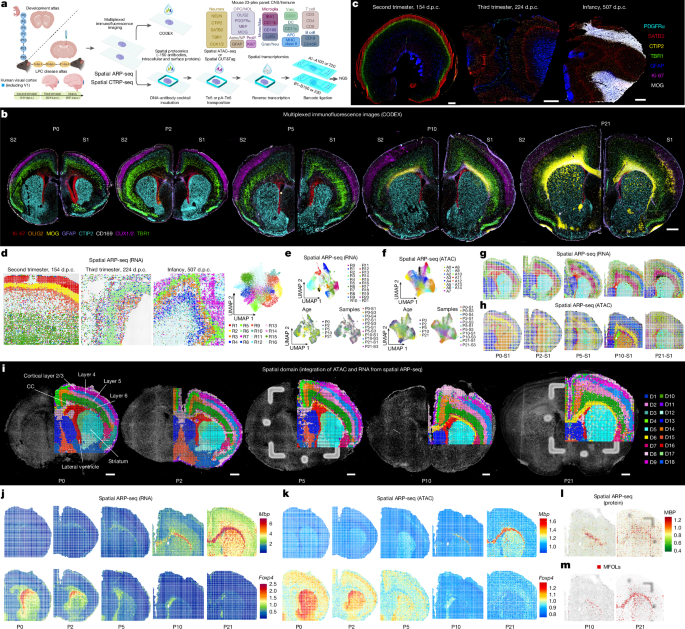
- Developed spatial tri-omics pipelines (spatial ARP‑seq and CTRP‑seq) to co-profile chromatin accessibility or H3K27me3, whole transcriptome and proteins in the same tissue pixel (15–20 μm resolution).
- CODEX multiplex imaging (mouse and human panels) provides protein-level single-cell spatial maps that benchmark tri-omic data across development.
- Chromatin accessibility for many cortical-layer TFs is spatially diffuse or temporally trailing relative to RNA — evidence of epigenetic persistence or priming of prior developmental states.
- Oligodendrocyte/myelin genes show early chromatin priming in the CC (P0–P5) before transcription and protein appear (P7–P21); myelination initiates laterally and progresses medially, with concurrent central and lateral programmes suggesting a bidirectional process.
- Projection neuron subtype maturation (CThPNs, SCPNs, CPNs) is spatially coordinated with oligodendrogenesis and myelination; axon tracts show RNA signatures likely reflecting transported transcripts.
- In an LPC demyelination model, CODEX and spatial tri-omics reveal lesion-core inflammation and a delayed, spatially distinct microglial/macrophage activation in distal white-matter tracts (e.g., stria medullaris, fornix); CD11c+ microglia dynamics and distinct microglial states (supportive vs inflammatory) are spatially and temporally resolved.
- Integration with single-cell atlases and benchmarking to human prenatal/infant samples shows cross-species conservation of major spatiotemporal programmes and technical feasibility for human tissue.
- All raw and processed data, plus an interactive portal, are publicly available (NeMO, GEO, Zenodo and spatial-omics.yale.edu).
Content summary
The authors combined high-plex protein imaging (CODEX) with DBiT-derived spatial ARP‑seq (ATAC–RNA–protein) and CTRP‑seq (CUT&Tag H3K27me3–RNA–protein) to obtain matched chromatin, transcriptional and protein maps at cellular resolution across mouse postnatal cortical and corpus callosum development and in a focal demyelination/remyelination model. They characterised layer-specific transcription factors, oligodendrocyte lineage progression and myelination timing, showing chromatin priming often precedes RNA and protein expression for myelin genes. Spatial regression models separated temporal, spatial and combined effects and produced integrated RNA/ATAC patterns, with GO enrichment linking clusters to neuronal maturation, synaptic transmission and oligodendrogenesis.
In the LPC model, multimodal spatial profiling and CODEX delineated lesion evolution (5, 10, 21 days post-lesion), revealed H3K27me3 changes associated with loss and recovery of oligodendrocyte markers, and exposed unexpected distal microglial activation propagating along white-matter tracts. Microglial subclusters exhibited distinct transcriptional and chromatin states (supportive vs pro-inflammatory), and ligand–receptor analyses suggested differing cell–cell interaction programmes between primary and distal lesion-like compartments.
The work is a resource: the authors provide data, code and an interactive browser to explore spatiotemporal tri-omic maps and support cross-species comparisons.
Context and relevance
This paper sits at the intersection of developmental neurobiology, epigenetics and neuroimmunology. It advances spatial multi-omics by measuring all three layers of the central dogma in situ and demonstrates how epigenetic priming, RNA expression and protein deposition are staggered across space and time during myelination and postnatal cortical maturation. For researchers studying myelination, oligodendrocyte heterogeneity, axon–glia interactions or MS-like lesions, the dataset and methods provide a high-value spatial reference and a technical blueprint for integrated epigenome–transcriptome–proteome studies. The distal propagation of microglial activation is especially relevant to models of spreading inflammation and may inform how focal lesions produce broader network-level immune responses.
Author style
Punchy: this is a heavyweight spatial‑omics paper and a practical atlas. If you work on cortical development, myelination, microglial states or spatial technologies, read the figures and methods — the tri‑omic workflows and the open datasets will save you time and open new experimental angles.
Why should I read this?
Short answer: because they didn’t just map RNA or proteins — they mapped chromatin, RNA and protein together in space. So if you want to know which genes are epigenetically primed, where myelination starts and how a focal lesion sends microglial ripples down white‑matter tracts, this paper gives you the maps and the data to dig in. Also — there’s an interactive portal, so you can go straight to the pixels that matter for your gene or cell type.