Transcriptomic and spatial organization of telencephalic GABAergic neurons
Summary
This Nature paper from the Allen Institute presents an integrated, high-resolution transcriptomic and spatial atlas of GABAergic neurons across the entire mouse telencephalon. Using >600k adult single-cell transcriptomes combined with MERFISH spatial data and developmental scRNA-seq across embryonic to early postnatal stages, the authors build a hierarchical taxonomy (classes → subclasses → supertypes → clusters) that maps cell types, their spatial distributions and inferred developmental origins.
The study highlights persistent transcription factor signatures that trace progenitor domains (MGE, CGE, LGE, POA, septum), reveals extensive long-range migration of related types, identifies spatial gene-expression gradients (for example across cortical layers and the striatum), and documents differing timing of diversification — cortical and striatal types diversify postnatally, whereas septal/pallidal/preoptic types reach mature profiles earlier.
Key Points
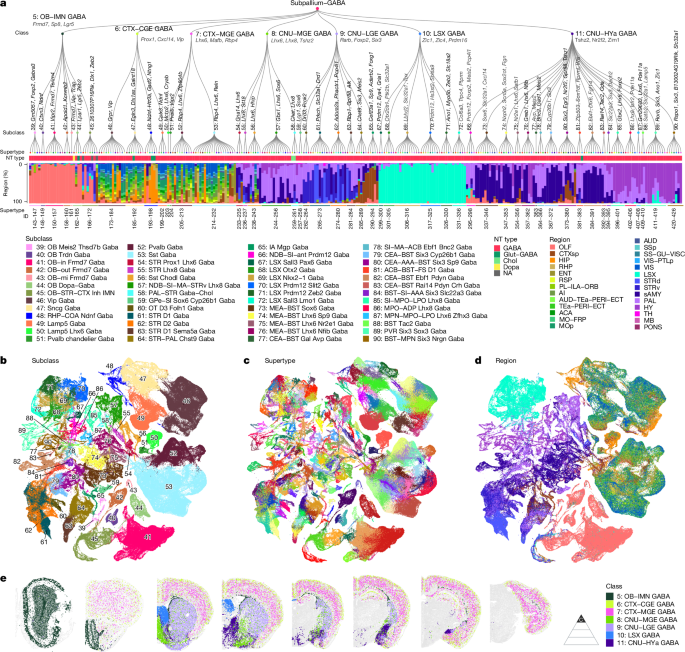
- Comprehensive taxonomy: 7 classes, 52 subclasses, 284 supertypes and 1,051 clusters for telencephalic GABAergic neurons, from >611,000 high-quality adult single-cell transcriptomes.
- Spatial mapping: MERFISH data registered to the Allen CCFv3 shows most supertypes span neighbouring regions; some (LSX, MOB/AOB) are region-restricted.
- Developmental tracing: integration of embryonic→postnatal scRNA-seq (614k developmental cells) links adult types to five principal progenitor domains (MGE, CGE, LGE, POA, septum) via persistent TF signatures.
- Long-range dispersion: many GABAergic supertypes occupy distant but connected brain nodes, reflecting wide migration from shared origins.
- Laminar and topographic gradients: MGE-derived Pvalb+ and Sst+ interneurons show depth-dependent gene gradients in cortex; D1/D2 MSNs show dorsolateral→ventromedial molecular gradients in striatum.
- Cell-type relations cross structures: examples include OB interneurons related to some amygdala/intercalated types and striatal MSN-like groups with distinct spatial patches.
- Cholinergic and co-release diversity: identification of cholinergic clusters (some co-releasing GABA or glutamate) across basal forebrain, striatum and septum.
- Timing of diversification differs: cortical and striatal classes continue to diversify postnatally (P0–P14 and beyond), whereas LSX and many CNU–HYa types mature largely prenatally.
- Resources released: processed data, MERFISH images and taxonomy are available via NeMO, BIL and the Allen Brain Cell Atlas for community use.
- Techniques and tools: combination of 10x single-cell RNA-seq, MERFISH spatial transcriptomics, scVI integration, ICA for gradients and BANKSY for spatial-domain clustering.
Context and relevance
This is a landmark resource for anyone working on inhibitory circuits, development of interneurons, basal ganglia organisation or spatial transcriptomics. It unifies transcriptomic identity, spatial location and developmental lineage at an unprecedented scale. The atlas will accelerate targeted experiments (genetic perturbations, connectivity and physiology studies) and offers candidate TF “terminal selectors” to test for maintenance of neuronal identity.
Why should I read this?
Short version: if you care about how inhibitory neurons are typed, where they live and where they came from, this paper is gold. It saves you months of digging through scattered datasets — the authors have done the heavy lifting and provide both the map and the markers you can use straightaway.
Author style
Punchy: big-team, big-data, big payoff. The paper isn’t just descriptive — it frames organisational principles (migration, gradients, timing) that change how we link development to adult circuit motifs. If your work touches circuits, development or molecular targeting, read the figures and the supplementary taxonomy.
Source
Article date: 05 November 2025