Convergent genome evolution shaped the emergence of terrestrial animals
Summary
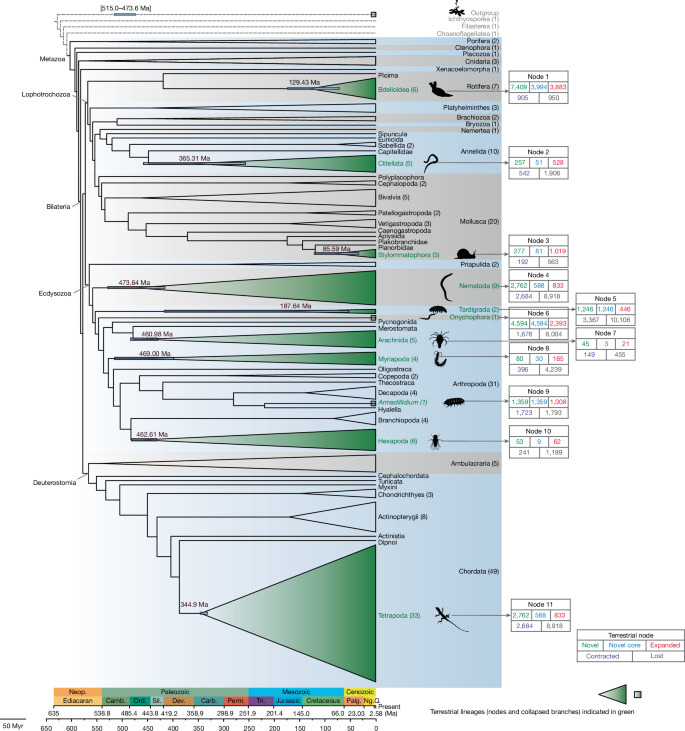
This Nature study uses a comparative genomics pipeline (InterEvo) on 154 genomes across 21 animal phyla to investigate how independent animal lineages moved from water onto land. The authors reconstruct ancestral homology groups (HGs) and identify patterns of gene gain, expansion, contraction and loss at 11 terrestrialisation nodes (from nematodes and arthropods to tetrapods and gastropods). They find repeated genomic changes — particularly gains in gene families linked to osmoregulation, stress response, immunity, sensory systems and metabolism — alongside notable gene reduction in other families. The study also defines three temporal windows when major terrestrial transitions occurred (Ordovician–Cambrian window, Devonian–Carboniferous, and Cretaceous) and distinguishes different genomic strategies in semi-terrestrial versus fully terrestrial lineages.
Key Points
- Large-scale gene turnover (novelty, expansions and losses) accompanies most independent transitions to land, signalling strong genome plasticity during terrestrialisation.
- Convergent gene gains repeatedly involve functions for osmoregulation, ionic transport, detoxification (cytochrome P450, flavin monooxygenases), stress resistance and sensory reception.
- Gene reductions (losses and contractions) are common and include convergent loss of some signalling domain families (e.g., Dbl‑homology and pleckstrin‑homology domains), suggesting shifts in regeneration/reproductive pathways.
- Semi‑terrestrial groups (water‑dependent) show broader, more versatile novel gene toolkits (cuticle remodelling, visual development, stress response), whereas fully terrestrial groups display a more streamlined set focused on neuronal development and membrane ion homeostasis.
- Arthropods and tetrapods show lineage‑specific adaptations: arthropods expand exoskeleton and visual genes; tetrapods enrich immune and epidermal defence genes linked to terrestrial skin barriers.
- The molecular timescale identifies three main windows of animal terrestrialisation that largely align with major plant and ecosystem changes on land.
Content summary
The authors clustered ~3.9 million protein sequences into 483,458 homology groups and reconstructed ancestral HG content at nodes representing 11 independent terrestrial transitions. They classified HGs as novel, novel core, expanded, contracted or lost and used permutation tests and the CAFE5 birth–death model to assess significance.
Most terrestrial events show elevated rates of novel gene emergence per million years compared with aquatic nodes (permutation test P = 0.0015). Functional annotation (GO and Pfam) of novel genes across events highlights shared functions: membrane ion transport, neurotransmitter/neuronal functions, lipid metabolism, detoxification and responses to stimuli.
Convergent expansions were detected in gene families for detoxification, oxidative stress and sensory reception (eg. cytochrome P450s, glutathione S‑transferases, GPCRs). Notable convergent losses include families involved in RhoGEF signalling domains. Semi‑terrestrial versus fully terrestrial comparisons reveal different enrichment patterns: the former retain diverse adaptive innovations to cope with intermittent moisture, the latter show narrower convergence likely reflecting more specialised, permanent terrestrial life.
A molecular clock based on 943 conserved orthologues places major animal terrestrialisation across three temporal windows: an early Cambrian–Ordovician cluster (arthropods, nematodes), a Devonian–Carboniferous cluster (clitellate annelids, tetrapods) and a Cretaceous cluster (bdelloid rotifers, land gastropods), linking evolutionary timing to changing terrestrial ecosystems.
Context and relevance
This paper provides a genome‑wide, cross‑phylum view of how animals repeatedly solved the challenges of life on land. The convergent emergence of functions for ion and water balance, detoxification and sensory adaptation shows that, despite different ancestries, similar ecological pressures can produce predictable molecular outcomes. The distinction between semi‑ and fully terrestrial genomic strategies is useful for evolutionary biologists, comparative genomics researchers and palaeobiologists seeking to link genomic innovation to fossil and environmental records. The InterEvo framework is also a methodological advance for detecting functional convergence across deep evolutionary time.
Why should I read this?
Want the big picture on how animals left the water without slogging through dozens of niche papers? This study does the heavy lifting: 154 genomes, 11 independent land invasions, and a clear list of the gene functions that kept popping up. If you care about evolution, genomes, or how life adapts to new worlds, this is a neat, data‑heavy map of the solutions evolution kept reaching for.
Author style
Punchy: the authors combine a rigorous pipeline with large taxon sampling to make a strong case that many genomic responses to terrestrial life are repeatable — but lineage‑specific twists matter. Read the details if you want the evidence and data behind those claims.