NSD2 targeting reverses plasticity and drug resistance in prostate cancer
Article Date: 26 November 2025
Article URL: https://www.nature.com/articles/s41586-025-09727-z
Article Image: Fig.2 (NSD2 / H3K36me2)
Summary
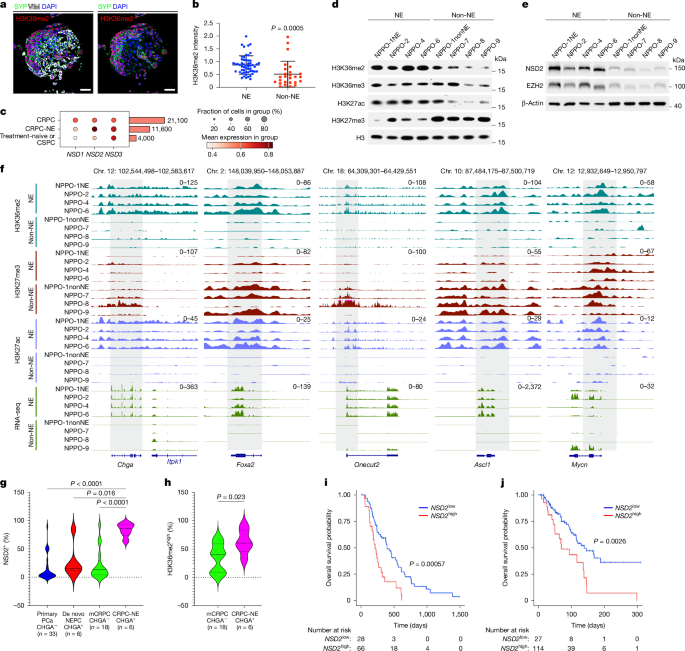
This study uses mouse and patient-derived organoid models to show that the histone methyltransferase NSD2 (MMSET/WHSC1) drives lineage plasticity in advanced prostate cancer and promotes resistance to androgen receptor (AR) inhibitors such as enzalutamide. Neuroendocrine-like, treatment-resistant castration-resistant prostate cancers (CRPC-NE) exhibit elevated H3K36me2 marks catalysed by NSD2. Genetic loss of NSD2 (CRISPR) or dominant-negative H3.3K36M reprogrammes the epigenome (loss of H3K36me2, gain of H3K27me3), restores AR expression and re-sensitises tumours to AR inhibition.
The authors synthesised a selective small-molecule NSD2 inhibitor (NSD2i; IC50 ≈ 3.8 nM against NSD2 on nucleosomes) and show that pharmacological NSD2 inhibition phenocopies genetic targeting: it reduces H3K36me2 domains, shifts cell states from neuroendocrine to adenocarcinoma-like programmes, synergises with enzalutamide to suppress growth and induces apoptosis in multiple CRPC organoid lines and xenografts. High NSD2 expression correlates with a neuroendocrine signature and worse overall survival in independent mCRPC cohorts, supporting translational relevance.
Key Points
- NSD2 and its product H3K36me2 are upregulated in CRPC-NE and in a subset of advanced tumours and correlate with poor patient survival.
- Organoid models (mouse NPp53-derived and human CRPC lines) reproduce the heterogeneity and transdifferentiation to neuroendocrine states observed in patients.
- Genetic targeting of NSD2 (CRISPR) or expression of H3.3K36M reduces H3K36me2, increases H3K27me3 and reverts neuroendocrine phenotype to AR+ adenocarcinoma-like states.
- A selective NSD2 inhibitor (NSD2i) reduces global H3K36me2 domains, reprogrammes enhancers tied to neuroendocrine regulators (ASCL1, FOXA2, ONECUT2) and shifts cell-state composition toward AR signalling.
- NSD2 inhibition alone has modest tumour effects, but combined pretreatment with NSD2i and enzalutamide shows strong synergy: reduced growth, increased apoptosis and loss of neuroendocrine markers in organoids and xenografts.
- NSD2 activity appears to antagonise PRC2/H3K27me3; targeting NSD2 reinstates repressive marks at neuroendocrine loci and disrupts plasticity programmes.
- Preclinical data provide a rationale for testing combined NSD2 and AR inhibition in advanced prostate cancer subtypes showing lineage plasticity or castration resistance.
Author style
Punchy — this is a heavyweight preclinical paper with clear translational promise. The work maps an epigenetic mechanism (NSD2/H3K36me2) for therapy-induced plasticity, demonstrates functional reversibility, and supplies a drug-like inhibitor that synergises with an approved AR antagonist. For clinicians and translational researchers this signals a plausible near-term therapeutic combination to test in resistant mCRPC.
Why should I read this?
Short version: if you care about why some prostate cancers stop responding to AR blockers, this paper tells you one clear mechanism and shows you a way to undo it. It’s a neat story — epigenetic change (NSD2 → H3K36me2) drives neuroendocrine transdifferentiation and drug resistance, and you can reverse that with a drug-like inhibitor so the tumour becomes sensitive to enzalutamide again. Saves you the reading time and gives a tangible therapeutic angle.
Context and relevance
Lineage plasticity is an important mechanism of resistance across solid tumours. This work positions NSD2 as a central epigenetic regulator of that plasticity in prostate cancer and links its activity to enhancer remodelling and expression of neural-specification regulators. Given an ongoing clinical effort to target NSD2-family enzymes, these data give strong preclinical support for combining NSD2 inhibition with AR-directed therapy in selected mCRPC patients — especially those with neuroendocrine features or high NSD2/H3K36me2 signatures.