Dated gene duplications elucidate the evolutionary assembly of eukaryotes
Summary
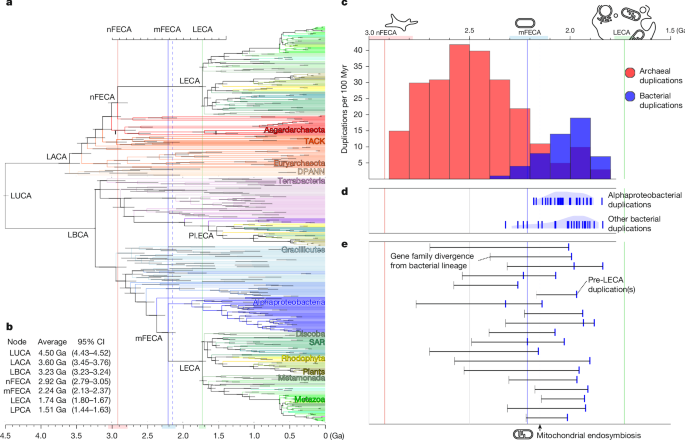
This Nature paper uses relaxed molecular clocks and a sequential Bayesian framework to date ancient gene duplications that occurred before the Last Eukaryotic Common Ancestor (LECA). By calibrating gene trees against a time-resolved species tree, the authors place the divergence of the nuclear stem (nFECA) at ~3.05–2.79 Ga, the alphaproteobacterial (mitochondrial) stem (mFECA) at ~2.37–2.13 Ga, and LECA at ~1.80–1.67 Ga. Key finding: many archaeal-origin duplications predate mitochondrial endosymbiosis, while a burst of alphaproteobacterial duplications around ~2.2 Ga marks mitochondrial integration. The data support a model in which a complex archaeal host (with cytoskeleton, endomembrane features and nascent nuclear systems) existed before mitochondrion acquisition — a “complexified archaeon, late-mitochondrion” (CALM) scenario.
Key Points
- Species-tree dating (MCMCTree) estimates nFECA at 3.05–2.79 Ga, mFECA at 2.37–2.13 Ga and LECA at 1.80–1.67 Ga.
- Archaeal-origin gene duplications are more numerous and began earlier than bacterial-origin duplications, suggesting major host complexification on the archaeal stem.
- Alphaproteobacterial-origin duplications cluster soon after mFECA, interpreted as evidence that mitochondrial endosymbiosis was already established by ~2.2 Ga.
- Core eukaryotic systems show staged duplication: cytoskeleton (actin/tubulin) and some endomembrane components duplicated early; endolysosomal specialisation and many bacterial-derived membrane functions came later.
- Spliceosomal components and RNA polymerase duplications indicate development of nucleus-related processes before mitochondrial acquisition.
- Preprotein import system and many mitochondrion-targeted paralogues date to ~2.3–2.0 Ga, constraining early mitochondrial integration.
- Results argue against “mitochondria-first” models; they favour models in which host complexity largely precedes mitochondrion acquisition (CALM models).
- The evolutionary timeline aligns mitochondrial acquisition roughly with the Great Oxidation Event (~2.4–2.2 Ga), situating eukaryogenesis in an important environmental transition.
Content summary
The study compiled 62 marker genes to build a cross-braced, time-resolved tree of life calibrated with 18 fossil constraints. Using a two-stage Bayesian approach, the dated species tree informed priors for single-gene relaxed-clock analyses (MCMCTree) to estimate ages for duplication nodes that predate LECA.
Of 135 well-resolved pre-LECA gene families analysed, 95 are of archaeal origin and 40 of bacterial origin. Duplication-age distributions show archaeal duplications beginning earlier (reflecting host genome elaboration), while alphaproteobacterial duplications appear rapidly after mFECA — interpreted as a fingerprint of mitochondrial integration. System-specific dating places branched actin (ARP2/3), tubulin paralogue expansions, early vesicle-trafficking duplications and spliceosomal/RNA polymerase duplications prior to or around the time of mitochondrial acquisition, whereas many bacterial-derived membrane and transporter duplications postdate it.
Context and relevance
This is a high-impact, methodologically rigorous attempt to order eukaryotic innovations in absolute time. By linking gene-duplication events to a dated species tree, the paper provides temporal constraints bearing directly on competing eukaryogenesis hypotheses (mitochondria-early vs mitochondria-late/intermediate). The conclusion that a complex archaeal host already possessed core cellular machinery before the mitochondrion arrived challenges energetic “mitochondria-first” narratives and supports models where host elaboration and endocytotic capability facilitated symbiosis.
It also ties biological events to Earth history: mitochondrial integration is placed close to the Great Oxidation Event, offering environmental context for a major evolutionary transition. The dataset and code are openly available and the authors performed extensive sensitivity tests to examine robustness to alternative topologies and calibrations.
Why should I read this?
Short, sharp: if you want to know which pieces of the eukaryotic cell came first and when — this paper does the heavy lifting. It dates duplication events that map to cytoskeleton, endomembrane, nucleus and mitochondrial systems and shows the host was already getting complicated before the mitochondrion rocked up. Read it if you care about how eukaryotes evolved, the timing of mitochondrial acquisition, or how molecular clocks can pin ancient cellular innovations to Earth history. In other words — saves you weeks of digging through technical gene-tree papers.