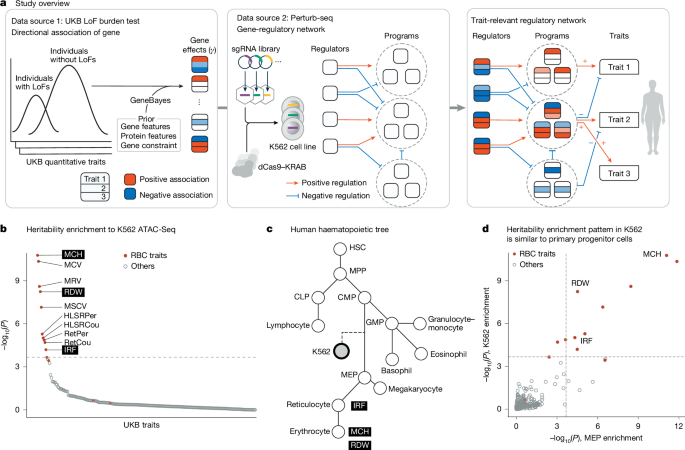
Causal modelling of gene effects from regulators to programs to traits
Article metadata
Article Date: 2025-12-10
Article URL: https://www.nature.com/articles/s41586-025-09866-3
Article Image: https://media.springernature.com/lw685/springer-static/image/art%3A10.1038%2Fs41586-025-09866-3/MediaObjects/41586_2025_9866_Fig1_HTML.png
Summary
This paper develops a systematic, genome-scale approach that links genes to cellular programmes and onward to organismal traits by combining loss-of-function (LoF) burden tests from large human cohorts with genome-wide Perturb-seq (CRISPR knockdown + single-cell RNA-seq). Using the K562 Perturb-seq dataset and UK Biobank/All of Us LoF data, the authors: (1) improve LoF effect estimates with an empirical‑Bayes method (GeneBayes); (2) identify 60 co‑expression “programmes” by consensus NMF; (3) measure regulator→gene and regulator→programme causal effects (β) from Perturb‑seq and correlate these with gene-level LoF effects (γ) to produce regulator–burden correlations; and (4) assemble compact gene→programme→trait graphs that explain and predict the direction of genetic effects for erythroid traits (MCH, RDW, IRF). They validate directional predictions with blood trans‑eQTLs and show when K562 is a suitable model (erythroid traits) and when trait‑matched perturbation data are needed.
Key Points
- Combines rare‑variant LoF burden tests with genome‑scale Perturb‑seq to trace causal regulatory paths from genes to traits.
- K562 Perturb‑seq data are particularly informative for erythroid traits (mean corpuscular haemoglobin (MCH), red cell distribution width (RDW), immature reticulocyte fraction (IRF)).
- GeneBayes (empirical Bayes) improves LoF effect (γ) estimates, increasing reproducibility and pathway enrichment.
- cNMF identified 60 programmes; perturbation effects on programme activity (β) allow regulator→programme mappings.
- Regulator–burden correlation (correlating β with γ across regulators) links perturbation effects to trait genetics and gives directionality.
- Constructed unified gene→programme→trait graphs that correctly predict the sign of many top gene effects and explain cross‑trait concordance/discordance.
- Validated programme directions using trans‑eQTLs and found both cell‑type specific and broadly shared programmes (eg cell‑growth programmes like MKI67).
- Model is generalisable but depends on having Perturb‑seq in trait‑relevant cell types; future expansion to more cell types, modalities and phenotypes is needed.
Context and relevance
Genome‑wide association studies and rare‑variant screens produce many gene‑trait links, but interpretation is hard because many associations act indirectly through regulatory networks. This work provides a practical framework to convert association signals into mechanistic, directional hypotheses by measuring causal effects from perturbations and integrating them with directional LoF estimates. It is directly relevant to researchers aiming to prioritise therapeutic targets, understand pleiotropy, or move from statistical hits to interpretable pathways.
Why should I read this?
Quick take: read this if you want a smart, experimentally grounded way to go from GWAS/LoF hits to mechanisms. The authors do the heavy lifting — merging CRISPR Perturb‑seq with rare‑variant effects and programme inference — so you don’t have to chase directionless enrichment lists. Expect clear maps that predict whether a gene increases or decreases a trait and explanations for oddball cases where traits diverge.
Author style
Punchy: the paper is practical and results‑driven. It doesn’t stop at enrichment; it builds predictive causal maps, validates them, and lays out the limitations (cell‑type matching, RNA readout limits). If you care about turning genetic signals into testable mechanisms or targets, the detail is worth your time.