Human assembloids recapitulate periportal liver tissue in vitro
Article Date: 17 December 2025
Article URL: https://www.nature.com/articles/s41586-025-09884-1
Article Image: https://media.springernature.com/lw685/springer-static/image/art%3A10.1038%2Fs41586-025-09884-1/MediaObjects/41586_2025_9884_Fig1_HTML.png
Summary
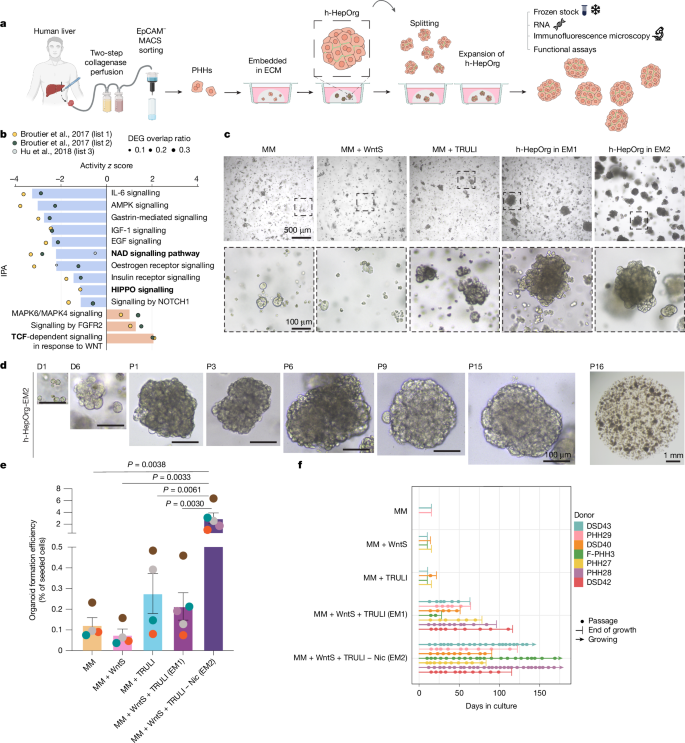
This Nature paper describes the generation of long-term expandable adult human hepatocyte organoids (h-HepOrgs) derived directly from patient liver tissue, and their combination with patient-matched cholangiocyte organoids and portal fibroblasts to create periportal “assembloids”. By activating WNT and YAP signalling (then reducing YAP for differentiation), the team established culture conditions (EM2 and DM) that permit serial expansion, cryopreservation and differentiation of hepatocyte organoids while preserving hepatocyte markers, bile canaliculi architecture and patient-specific transcriptional signatures. Assembloids self-organise into structures that mirror periportal architecture, show portal-like hepatocyte functions (urea synthesis, gluconeogenesis) and — when mesenchyme is increased — model hallmarks of biliary fibrosis. The work includes single-cell and bulk RNA-seq benchmarking against human liver atlases, functional assays, drug metabolism measurements and xenotransplantation tests that demonstrate engraftment and rescue capacity in mice.
Key Points
- Established EM2 medium (WNT surrogate + LATS inhibitor without nicotinamide) enables long-term expansion (>10 passages) and cryopreservation of adult human hepatocyte organoids from 28 donors with 100% efficiency.
- Transient activation of YAP and WNT drives proliferation; switching to a differentiation medium (DM) reduces YAP activity and promotes mature hepatocyte traits and improved bile canaliculi morphology.
- h-HepOrgs retain patient-specific gene-expression signatures and inter-donor variation in bile canaliculi architecture and drug metabolism (e.g. verapamil → norverapamil), supporting personalised modelling.
- Combining h-HepOrgs with patient-derived cholangiocyte organoids and CD90+PDGFRA+ portal fibroblasts yields periportal assembloids that self-organise into tissue-like arrangements with duct-like lumina in close contact with mesenchyme.
- Single-cell RNA-seq shows assembloid cell types correlate strongly with in vivo liver atlas populations; hepatocytes in assembloids adopt a more periportal transcriptional identity and show enhanced portal functions (urea, gluconeogenesis).
- Increasing mesenchymal cell numbers produces “fibrotic-like” assembloids that reproduce transcriptional and morphological features of biliary fibrosis (inflammation, ECM/matrix programmes, cholangiocyte expansion, hepatocyte apoptosis).
- Expanded and differentiated h-HepOrgs engraft in Fah−/− Rag2−/− Il2rg−/− mice and can rescue the lethal phenotype, demonstrating functional competence in vivo.
- Resources (bulk and scRNA-seq data, code and a living biobank) are deposited under controlled access to protect donor privacy; organoid lines available under material transfer agreement.
Content summary
The authors overcame previous limitations in human hepatocyte culture by combining a refined isolation protocol (EpCAM MACS separation) with pathway-guided media design. IPA of regenerative and cancer datasets highlighted WNT and YAP as drivers; activating these pathways (Wnt surrogate + TRULI) allowed expansion, while nicotinamide removal improved longevity. Differentiation (DM) restored many mature hepatocyte functions and polarity, producing elongated, interconnected bile canaliculi similar to tissue. By isolating and expanding portal fibroblasts (CD90+PDGFRA+) and labelling cholangiocytes and fibroblasts with nuclear fluorescent markers, they optimised ratios and aggregation conditions (AggreWell) to form reproducible periportal assembloids. Functional testing showed assembloids outperform isolated hepatocyte organoids for periportal functions and capture patient-specific metabolic phenotypes. Doubling down on mesenchyme reproduced features of biliary fibrosis, including inflammatory signalling and ECM deposition signatures. Extensive sequencing and comparison to human liver atlases support the physiological relevance of the model.
Context and relevance
This study addresses a major gap: faithful human periportal tissue models that capture epithelial–stromal interactions, patient heterogeneity and disease-relevant responses. Existing systems either lack long-term adult hepatocyte expansion, three-dimensional bile canaliculi structure, stromal components or patient specificity. The assembloid platform enables mechanistic studies of portal biology, personalised toxicology and drug metabolism assays, and offers a tractable system for modelling cholestatic diseases (PSC, PBC) and fibrogenesis without immediate reliance on animal models. For translational research, the living biobank and engraftment data hint at future uses in precision medicine and cell therapy development—though incorporation of vasculature and immune components will be needed to model all periportal triad aspects.
Why should I read this?
Because if you work on liver biology, drug metabolism, cholangiopathies or organoid-based disease modelling, this paper saves you months of bootstrapping. It shows a reliable recipe for growing adult human hepatocyte organoids, how to slot in cholangiocytes and portal fibroblasts to get periportal architecture, and gives a ready-made platform for testing fibrosis cues or drug metabolism with patient-specific readouts. In short: they did the hard work so you don’t have to — and the assembloids actually behave like human periportal tissue.
Author style
Punchy: the team demonstrate a clear step-change — from non-expanding hepatocyte preps to a robust, patient-derived organoid and assembloid platform that recapitulates periportal structure and function. This is highly relevant to anyone aiming to model human liver disease, toxicity or personalise drug testing: the methods and biobank materially advance what can be modelled in vitro.