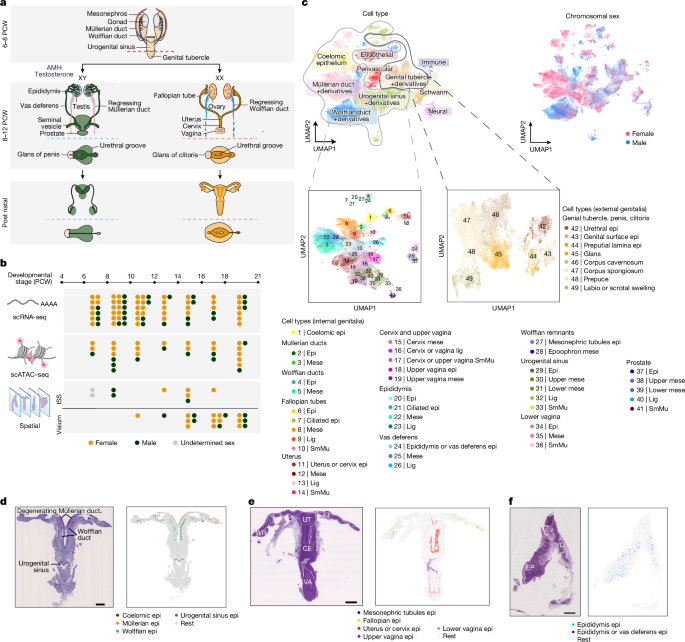
Spatiotemporal cellular map of the developing human reproductive tract
Summary
This Nature paper presents a comprehensive, multi-omic, spatiotemporal atlas of the developing human reproductive tract (excluding gonads), profiling >500,000 single cells from 89 fetal samples spanning 6–21 post-conceptional weeks (PCW). Using scRNA-seq, scATAC–seq, in situ sequencing (ISS) and 10x Visium spatial transcriptomics, the authors define 52 reproductive-tract-specific cell types, map Müllerian and Wolffian duct emergence, patterning and regression, identify key transcriptional regulators and cell–cell communication pathways that drive regionalisation, and predict susceptibilities to clinically approved drugs and endocrine-disrupting chemicals (EDCs). They validate EDC effects (BPA and BBP) on fetal-derived uterine organoids and make the dataset available via a public portal.
Key Points
- Large multi-omic atlas: 538,742 scRNA-seq cells, 226,668 scATAC cells and >1.8M ISS-resolved cells across 6–21 PCW, integrated with Visium spatial data.
- Identified 52 specialised cell types across internal and external genitalia; spatial mapping was essential to annotate previously undescribed markers.
- Resolved Müllerian duct emergence, migration and male-specific regression pathways — including migratory gene programmes in epithelium/mesenchyme and male upregulation of WNT inhibitors and autophagy markers.
- Refined HOX-code and discovered additional spatially variable transcription factors that define rostrocaudal regionalisation of Müllerian and Wolffian ducts (fallopian tube ⇢ uterus ⇢ cervix ⇢ vagina; epididymis ⇢ vas deferens).
- Mapped mesenchymal–epithelial signalling (WNT, retinoic acid, BMP, IGF, integrins, RSPO/LGR5) that instructs epithelial identity and regional differentiation; some gradients persist into adulthood.
- Characterised sexual dimorphism in the genital tubercle during the masculinisation programming window, pinpointing androgen-responsive mesenchymal and epithelial genes and candidate pathways (Notch, adherens junction molecules) implicated in urethral canalisation.
- Predicted cell-type vulnerabilities to 47 clinically used drugs and several EDCs; experimentally validated that BPA and BBP shift fetal uterine organoid cell composition (increased ciliogenesis) and induce oestrogen-responsive gene programmes.
- All raw data, imaging and code openly available (ArrayExpress accessions and GitHub repository) and the interactive atlas is at reproductivecellatlas.org.
Content Summary
The authors collected 89 human fetal reproductive-tract samples (6–21 PCW), applying scRNA-seq, scATAC–seq, ISS and 10x Visium. Integrating dissociated single-cell profiles with spatial data enabled high-confidence annotation of 52 cell types across the internal and external genitalia and neighbouring tissues. Early (≤10 PCW) samples reveal coexisting Müllerian and Wolffian ducts and the coelomic epithelium origins; later stages show clear female- and male-specific organ identities.
Detailed trajectory and smFISH analyses reconstruct Müllerian duct emergence from coelomic epithelium, migratory gene programmes and a male-specific degenerating mesenchymal branch marked by WNT inhibitors (NOTUM, NKD1, WIF1) and autophagy markers. Using a computational rostrocaudal axis, the study maps HOX and other transcription-factor gradients that regionalise ducts into fallopian tube, uterus, cervix, upper vagina and the male analogues in the Wolffian axis. Mesenchymal ligands (WNT4/5A, ALDH1A1) and inhibitors (WIF1, SFRP5), BMPs and integrin ligands show spatially restricted expression with corresponding epithelial receptor patterns.
In the external genitalia, no wholly sex-specific cell types were found, but sex-biased gene expression in the early corpus spongiosum and urethral epithelium points to androgen-driven programmes (e.g., CSRP2, SRD5A2, SCGB1A1, PTPRD) and candidate cell–cell interactions (JAG1–NOTCH2/3) for urethral canalisation. The atlas was used to predict drug/EDC targets and windows of susceptibility; organoid exposures to BPA and BBP corroborated oestrogen-like effects and increased ciliogenesis, validating the approach.
Context and relevance
This resource fills a major gap: a holistic, cell-resolved map of human reproductive-tract development across the key windows of sexual differentiation. It moves beyond rodent inference by providing human-specific spatial and regulatory detail — critical for linking genetic variants to developmental timing and cell types, improving in vitro modelling, and informing toxicology and drug-safety assessments for gestational exposures.
Author style
Punchy: This is a big deal. The paper supplies a meticulously integrated, openly available atlas that will be a reference for developmental biologists, reproductive medicine researchers, toxicologists and anyone modelling human reproductive tissues. The combination of multi-omics, spatial mapping and experimental validation makes the dataset immediately useful for prioritising candidate genes, regulatory elements and vulnerable developmental windows.
Why should I read this?
Quick and blunt — read this if you care about how human reproductive organs form, why congenital anomalies or later-life reproductive diseases happen, or how drugs and everyday chemicals might mess with foetal development. It’s packed with human-specific maps, candidate genes and validated EDC effects, plus the data and code are public so you can dig straight in without repeating the heavy lifting.