RNA-triggered Cas12a3 cleaves tRNA tails to execute bacterial immunity
Summary
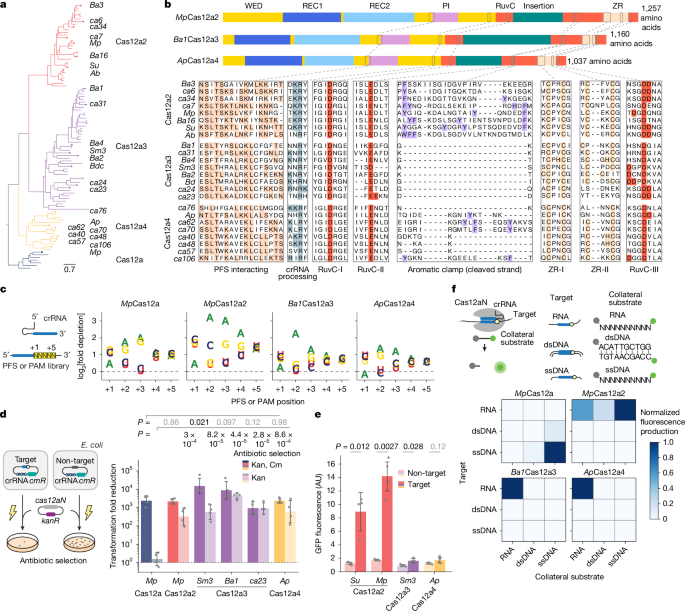
Researchers report a new clade of CRISPR-associated nucleases, dubbed Cas12a3, that are activated by complementary target RNA and then selectively trim the universally conserved 3′ CCA tails of tRNAs. Rather than cleaving the recognised target RNA or damaging DNA, activated Cas12a3 loads free tRNAs via a specialised tRNA-loading domain (tRLD) into its RuvC nuclease and excises the tRNA 3′ tail. Widespread trimming of tRNA tails disrupts translation, triggers growth arrest and limits phage spread. High-resolution cryo-EM structures reveal how Cas12a3 recognises the crRNA:target duplex and positions the tRNA acceptor stem and CCA tail for precise cleavage. The authors also exploit Cas12a3’s unique substrate preference to build an orthogonal reporter for multiplexed RNA detection alongside Cas13 systems.
Key Points
- Discovery of Cas12a3: a distinct Cas12a-related clade (Cas12a2, Cas12a3, Cas12a4) found in environmental metagenomes that is RNA-activated.
- Substrate specificity: after target RNA recognition, Cas12a3 preferentially cleaves the conserved 3′ CCA tail of tRNAs rather than extensively cleaving the recognised target RNA or cellular DNA.
- Mechanism: a unique tRNA-loading domain (tRLD) and REC2 interactions position the tRNA acceptor stem and CCA tail into the RuvC active site for trimming.
- Biological effect: broad trimming of tRNA 3′ tails disables translation, causing growth arrest and preventing phage dissemination without detectable DNA damage or reliance on the RelA stringent-response pathway.
- Structural evidence: cryo-EM structures (binary, ternary, quaternary, post-cleavage) map conformational changes from activation to tRNA capture and show the cleaved ACCA fragment wedged between RuvC and tRLD.
- Application: engineered short structured reporters that mimic tRNA tails enable Cas12a3 to be combined with Cas13 enzymes for multiplexed, one-pot RNA detection (e.g. SARS-CoV-2, RSV, influenza A) with orthogonal readouts.
- Diversity & evolution: Cas12a3 differs functionally from Cas12a2 (broad collateral cleavage) and Cas13 (target-centred cleavage), illustrating functional diversification within Cas12-family enzymes.
Context and relevance
Cleavage or inactivation of tRNAs is an increasingly recognised antiviral strategy across life. Cas12a3 adds a CRISPR-based example that targets the universally conserved tRNA tail (CCA), a site that phages cannot easily evade by simple anticodon mutations. The structural and biochemical characterisation explains substrate selectivity and suggests Cas12a3 is a naturally evolved translational-silencing defence distinct from DNA-targeting and broad collateral-RNase strategies. For researchers working on phage resistance, RNA-targeting CRISPR tools or diagnostics, Cas12a3 both expands mechanistic understanding of prokaryotic immunity and offers a new programmable tool with a unique substrate preference.
Why should I read this?
Because it’s not just another Cas nuclease paper. These folks found a CRISPR enzyme that, when told to look at an invader RNA, goes off and chops off tRNA tails across the cell instead of shredding DNA. That’s a clever, hard-to-escape strategy and gives you a new lever for diagnostics and maybe for targeted translational arrest. Quick read if you want the guts of a new CRISPR mechanism and a neat applied trick for multiplexed RNA detection.