An expanded registry of candidate cis-regulatory elements
Article Date: 07 January 2026
Article URL: https://www.nature.com/articles/s41586-025-09909-9
Summary
The ENCODE consortium has released an expanded, functionally annotated registry of candidate cis-regulatory elements (cCREs). The new ENCODE4 registry comprises ~2.37 million human and ~0.97 million mouse cCREs, covering roughly 21% of the human genome and substantially increasing biosample coverage. The update integrates large-scale biochemical maps, transcription factor ChIP–seq and ATAC/DNase data plus high-throughput functional assays (STARR-seq, MPRAs, CRISPR perturbations and transgenic assays). Crucially, more than 97% of human cCREs have been assayed in at least one functional experiment and roughly 28% showed significant activity in one or more tests. The resource identifies new element classes (CA-TF, CA and TF cCREs), systematic silencer subclasses (including REST-bound silencers and STARR-defined silencers), and marks latent, stimulus-responsive enhancers bound by MAFF/MAFK. The registry is accessible via the SCREEN portal and is illustrated with a detailed dissection of an RBC-trait locus that nominates KLF1 as a likely causal gene.
Key Points
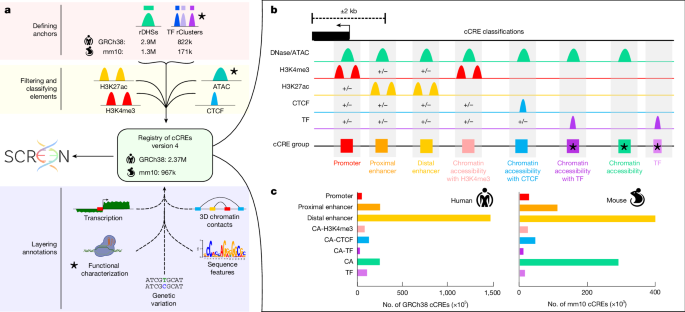
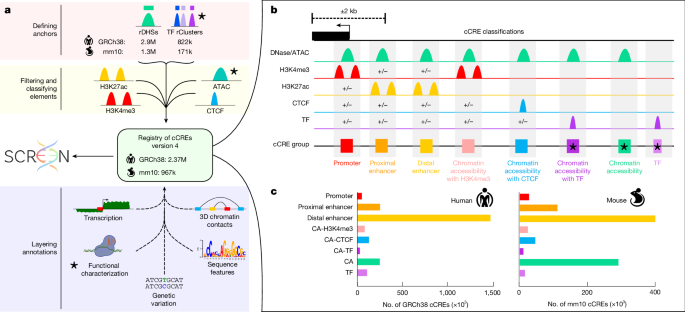
- ENCODE4 expands the cCRE catalogue to ~2.37M human and ~926k mouse elements (threefold increase over ENCODE3), spanning 21% of the human genome.
- The registry integrates biochemical signatures, TF binding, chromatin accessibility and large-scale functional assays; 97% of human cCREs were covered by at least one functional assay.
- Around 28% of tested cCREs show significant activity in at least one assay; STARR-seq (with CAPRA analysis) enabled high-throughput enhancer and silencer scoring across millions of fragments.
- New cCRE classes were added (CA-TF, CA and TF), which capture elements with low accessibility or TF-only binding and reveal silencers and latent/dynamic enhancers.
- Silencers were systematically annotated: REST-bound classes (dual-function enhancer/silencers and exclusive silencers) and STARR-defined silencers (stringent and robust) total ~9,972 silencer cCREs.
- MAFF/MAFK-bound cCREs mark a class of latent, stimulus-responsive enhancers poised for activation under stress or specific stimuli.
- The SCREEN web portal provides access to the registry and layered ENCODE annotations for integrative analyses and post-GWAS interpretation.
- Practical application: analysis of the RTBDN–MAST1 RBC-trait locus prioritised KLF1 (and PRDX2 as a secondary candidate) and nominated specific variants for functional follow-up.
Content summary
The paper describes the pipeline improvements and additional data that underlie ENCODE4: expanded DNase/ATAC and ChIP–seq experiments, incorporation of thousands of TF ChIP datasets, improvements to recover regions in duplicated or repetitive loci, and expanded core biosample coverage (170 core samples with the four core assays). cCREs were classified into eight biochemical classes using accessibility and histone/CTCF marks; three new categories (CA-TF, CA and TF) broaden the registry to elements that would be missed by accessibility-only approaches. The authors introduce CAPRA (CRE-centric analysis and prediction of reporter assays) to assign STARR-seq activity scores to individual cCREs within larger fragments; CAPRA enables detection of both enhancers and silencers and quantifies combinatorial interactions between adjacent cCREs.
Functional assays revealed cell-type-specific activity patterns (promoters are more consistent across cell types than distal enhancers), motif enrichments that explain activity differences (for example, GATA1 in K562, HNF4A and p53 in HepG2), and cooperative or repressive interactions among nearby elements. REST ChIP–seq was used to define REST+ cCREs and to separate REST+ enhancer/silencer elements (context-dependent) from REST+ silencers (constitutively repressive outside neuronal contexts). STARR-seq-based negative scores identified additional silencers, many overlapping LINE elements and showing reduced expression of nearby genes. MAFF/MAFK-bound elements appear poised for activation and are enriched near development and stress-response genes. Finally, the authors demonstrate how the registry aids variant-to-gene mapping and prioritisation at an RBC-trait locus, nominating KLF1 as the top candidate and highlighting variants for follow-up.
Context and relevance
This expanded registry is one of the most comprehensive reference maps of putative regulatory elements to date. It matters because non-coding regulatory variation underpins much of human trait and disease biology: having a broad, functionally annotated catalogue accelerates interpretation of GWAS hits, variant prioritisation, and experimental design for mechanistic studies. The addition of silencers and TF-only classes addresses recognised blind spots in prior atlases that focused on open chromatin and active histone marks. The combined biochemical, sequence and functional data make the resource especially useful for computational model training, variant annotation pipelines, and choosing candidate regions for CRISPR perturbation or reporter validation.
Why should I read this
Want the short version? ENCODE just dropped a huge, usable map of regulatory DNA — with activity data — so you don’t have to trawl disparate datasets. If you work on gene regulation, GWAS follow-up, variant interpretation or building predictive models, this will save you weeks of data wrangling and give better targets to test.